Probe CUST_25661_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25661_PI426222305 | JHI_St_60k_v1 | DMT400037378 | TAAGCTTACTGACCTTCGTTCTGCCAATGTAAGTACGGAAAAGCTTGCTAGAGTTGCCGT |
All Microarray Probes Designed to Gene DMG400014420
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25661_PI426222305 | JHI_St_60k_v1 | DMT400037378 | TAAGCTTACTGACCTTCGTTCTGCCAATGTAAGTACGGAAAAGCTTGCTAGAGTTGCCGT |
Microarray Signals from CUST_25661_PI426222305
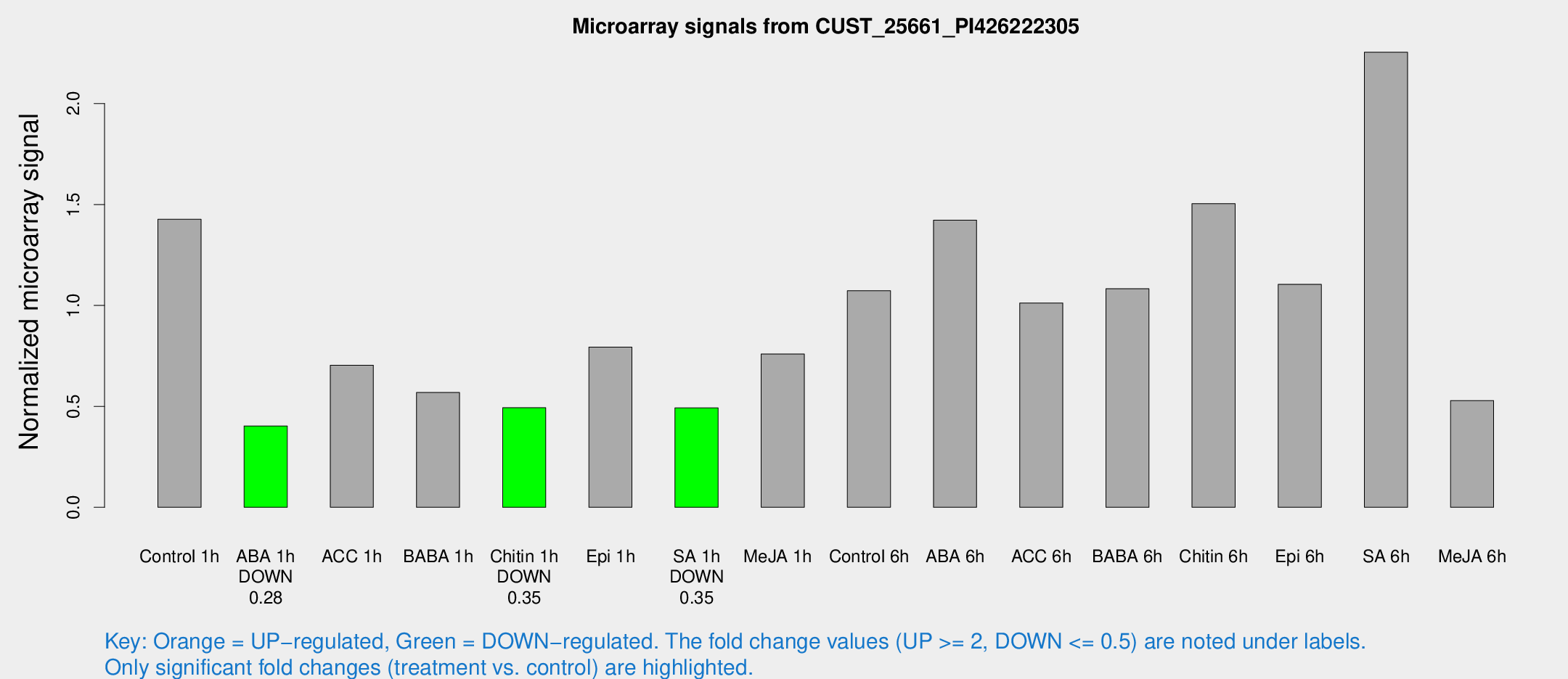
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 25.5061 | 3.35598 | 1.42714 | 0.188039 |
| ABA 1h | 6.47893 | 2.95833 | 0.403132 | 0.190261 |
| ACC 1h | 13.6185 | 3.3871 | 0.703923 | 0.201824 |
| BABA 1h | 10.9616 | 3.58778 | 0.568744 | 0.227425 |
| Chitin 1h | 8.18208 | 3.11192 | 0.493166 | 0.204282 |
| Epi 1h | 13.1623 | 3.16104 | 0.793647 | 0.261782 |
| SA 1h | 9.1348 | 3.0959 | 0.492674 | 0.171891 |
| Me-JA 1h | 12.477 | 3.69201 | 0.759369 | 0.313535 |
| Control 6h | 24.5039 | 9.65726 | 1.07304 | 0.584663 |
| ABA 6h | 28.3785 | 5.7728 | 1.42237 | 0.27597 |
| ACC 6h | 21.1419 | 4.07528 | 1.01218 | 0.302705 |
| BABA 6h | 25.9688 | 10.0217 | 1.08254 | 0.507496 |
| Chitin 6h | 29.5238 | 4.97078 | 1.5042 | 0.297354 |
| Epi 6h | 24.169 | 6.5314 | 1.10469 | 0.42149 |
| SA 6h | 66.8506 | 42.742 | 2.25427 | 2.93359 |
| Me-JA 6h | 12.932 | 7.47025 | 0.528906 | 0.300794 |
Source Transcript PGSC0003DMT400037378 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G20420.1 | +1 | 1e-84 | 263 | 130/252 (52%) | RNAse THREE-like protein 2 | chr3:7119371-7120895 REVERSE LENGTH=391 |
