Probe CUST_25638_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25638_PI426222305 | JHI_St_60k_v1 | DMT400029318 | GGGACTGGAATTTGTAACTTAGCCCAGCTTGAATGTTTGACATTGATTCCTAATAATATT |
All Microarray Probes Designed to Gene DMG400011266
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25638_PI426222305 | JHI_St_60k_v1 | DMT400029318 | GGGACTGGAATTTGTAACTTAGCCCAGCTTGAATGTTTGACATTGATTCCTAATAATATT |
Microarray Signals from CUST_25638_PI426222305
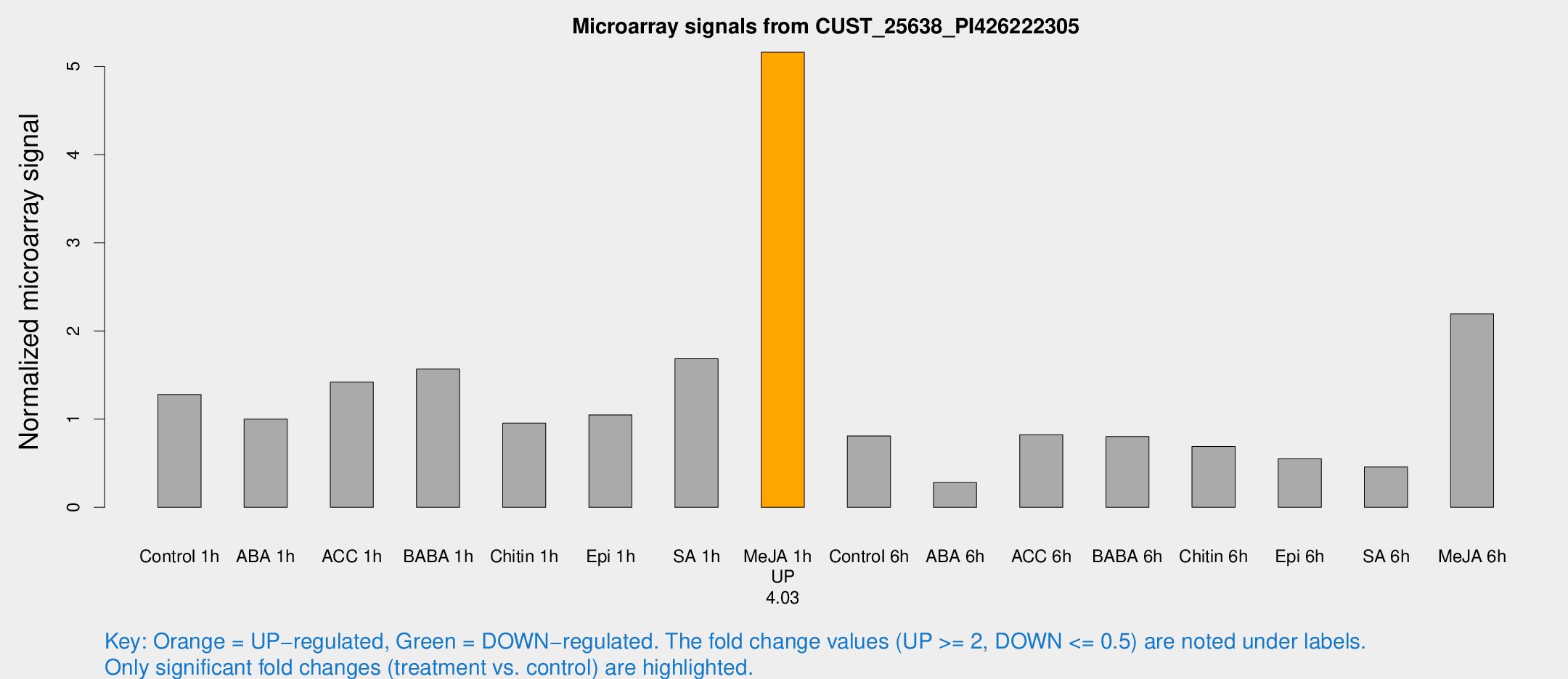
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 4016.84 | 961.11 | 1.27939 | 0.215774 |
| ABA 1h | 2807.67 | 709.114 | 1.00045 | 0.182772 |
| ACC 1h | 4790.84 | 1546.08 | 1.42079 | 0.39494 |
| BABA 1h | 4514.38 | 261.112 | 1.56851 | 0.101559 |
| Chitin 1h | 2782.51 | 830.654 | 0.955065 | 0.276305 |
| Epi 1h | 2747.97 | 353.128 | 1.04803 | 0.112136 |
| SA 1h | 6319.46 | 2802.87 | 1.68455 | 0.970221 |
| Me-JA 1h | 12680.9 | 1207.3 | 5.16135 | 0.364731 |
| Control 6h | 2939.94 | 1290.91 | 0.80852 | 0.322965 |
| ABA 6h | 972.162 | 290.924 | 0.279733 | 0.102606 |
| ACC 6h | 2995.12 | 675.392 | 0.822397 | 0.199266 |
| BABA 6h | 2793.1 | 532.727 | 0.802081 | 0.183576 |
| Chitin 6h | 2250.97 | 381.162 | 0.690551 | 0.111619 |
| Epi 6h | 1909.2 | 326.227 | 0.549963 | 0.137537 |
| SA 6h | 1455.55 | 419.542 | 0.457815 | 0.104985 |
| Me-JA 6h | 6598.79 | 738.565 | 2.19335 | 0.419879 |
Source Transcript PGSC0003DMT400029318 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G54160.1 | +3 | 3e-128 | 380 | 200/364 (55%) | O-methyltransferase 1 | chr5:21982075-21984167 FORWARD LENGTH=363 |
