Probe CUST_25623_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25623_PI426222305 | JHI_St_60k_v1 | DMT400029242 | CGTCTTTTCACAAGGATGTAATTTTATACACTTGCTACTATGGAGAGGTGGCAATTTTTT |
All Microarray Probes Designed to Gene DMG400011246
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25623_PI426222305 | JHI_St_60k_v1 | DMT400029242 | CGTCTTTTCACAAGGATGTAATTTTATACACTTGCTACTATGGAGAGGTGGCAATTTTTT |
Microarray Signals from CUST_25623_PI426222305
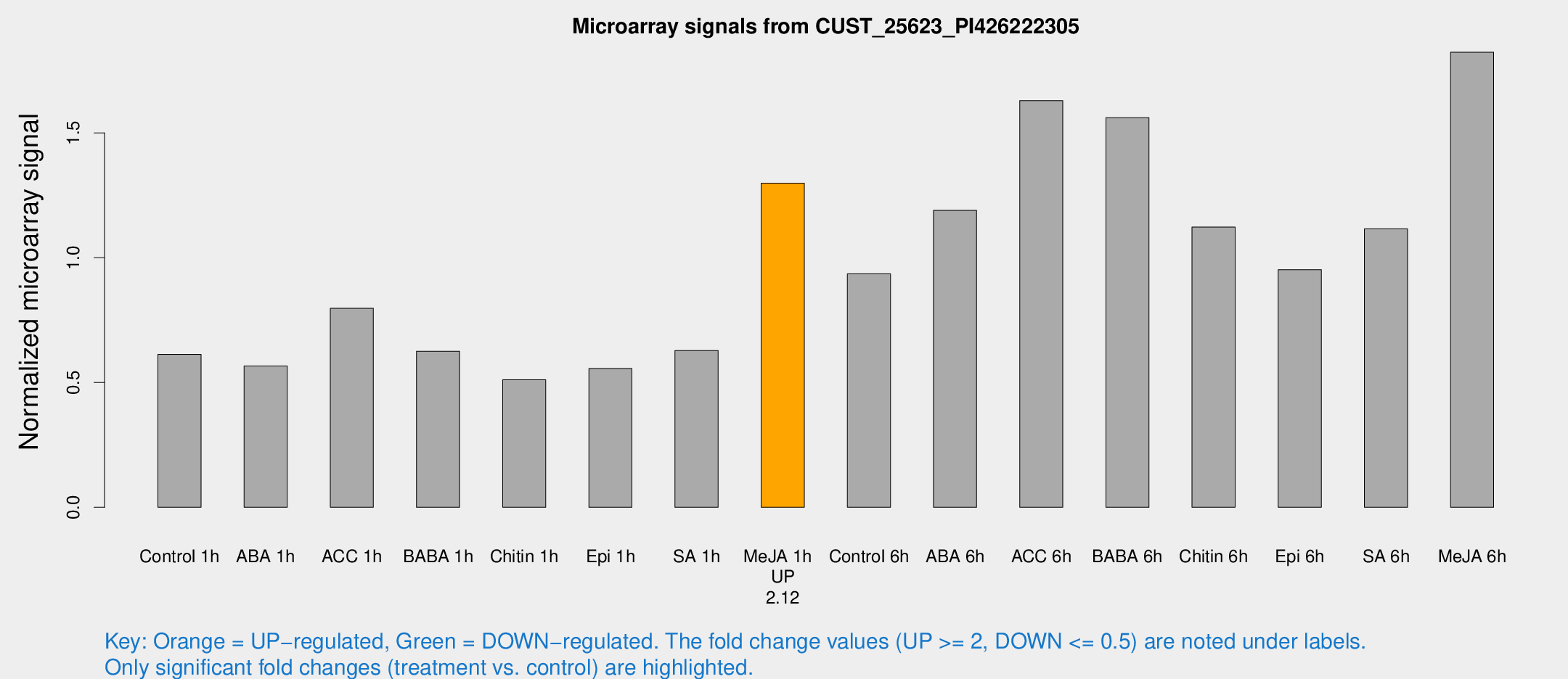
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 855.6 | 96.6965 | 0.612513 | 0.036973 |
| ABA 1h | 707.008 | 106.008 | 0.56596 | 0.0469838 |
| ACC 1h | 1208.25 | 297.375 | 0.797192 | 0.16391 |
| BABA 1h | 834.548 | 48.4578 | 0.625301 | 0.0361961 |
| Chitin 1h | 646.262 | 85.9478 | 0.510774 | 0.0515124 |
| Epi 1h | 670.655 | 65.9742 | 0.555667 | 0.0467509 |
| SA 1h | 938.276 | 222.294 | 0.62822 | 0.141069 |
| Me-JA 1h | 1486.92 | 181.462 | 1.29835 | 0.0750233 |
| Control 6h | 1461.19 | 427.66 | 0.934913 | 0.280963 |
| ABA 6h | 1759.08 | 156.029 | 1.1898 | 0.0687382 |
| ACC 6h | 2610.06 | 291.324 | 1.62919 | 0.0940956 |
| BABA 6h | 2429.73 | 214.037 | 1.56096 | 0.0901581 |
| Chitin 6h | 1649.49 | 95.3446 | 1.12232 | 0.0648504 |
| Epi 6h | 1502.04 | 180.439 | 0.951987 | 0.110169 |
| SA 6h | 1548.67 | 207.299 | 1.11554 | 0.145009 |
| Me-JA 6h | 2528.2 | 240.561 | 1.82294 | 0.155874 |
Source Transcript PGSC0003DMT400029242 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G79530.1 | +3 | 0.0 | 569 | 336/426 (79%) | glyceraldehyde-3-phosphate dehydrogenase of plastid 1 | chr1:29916232-29919088 REVERSE LENGTH=422 |
