Probe CUST_2550_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2550_PI426222305 | JHI_St_60k_v1 | DMT400072450 | TCCAACAAAAGGGTGTTCTTGAAGTTCCTTGTAGCGCCGCGGAATTGGAAAAGATTTTGG |
All Microarray Probes Designed to Gene DMG400028186
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_2572_PI426222305 | JHI_St_60k_v1 | DMT400072451 | GATTTTGGAAATCTAAGATTGGATATTAAGATAGTTCCTTCTATGCTAGCTAGAGCTAGG |
| CUST_2550_PI426222305 | JHI_St_60k_v1 | DMT400072450 | TCCAACAAAAGGGTGTTCTTGAAGTTCCTTGTAGCGCCGCGGAATTGGAAAAGATTTTGG |
Microarray Signals from CUST_2550_PI426222305
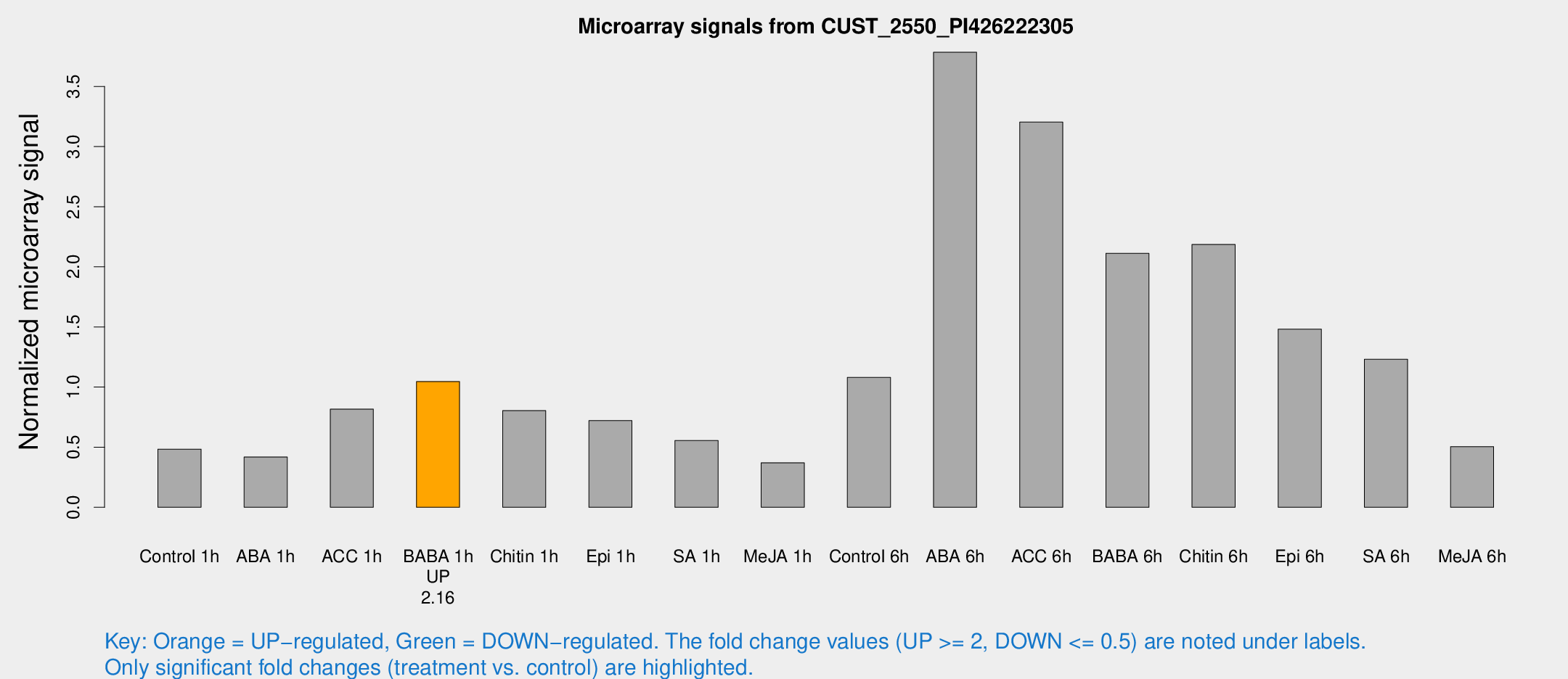
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 113.987 | 21.7756 | 0.483793 | 0.0825239 |
| ABA 1h | 97.9038 | 31.6977 | 0.41834 | 0.175377 |
| ACC 1h | 226.399 | 76.8568 | 0.816723 | 0.3431 |
| BABA 1h | 233.364 | 33.3705 | 1.04572 | 0.102665 |
| Chitin 1h | 164.788 | 10.5609 | 0.803719 | 0.103917 |
| Epi 1h | 148.758 | 32.7382 | 0.72071 | 0.164381 |
| SA 1h | 129.563 | 8.09527 | 0.554935 | 0.0446675 |
| Me-JA 1h | 69.2185 | 6.7036 | 0.370056 | 0.027376 |
| Control 6h | 329.934 | 137.235 | 1.07981 | 0.695105 |
| ABA 6h | 1164.68 | 594.762 | 3.7843 | 2.47076 |
| ACC 6h | 935.801 | 332.293 | 3.20361 | 0.655998 |
| BABA 6h | 614.484 | 236.64 | 2.112 | 0.758018 |
| Chitin 6h | 556.711 | 127.622 | 2.18593 | 0.559815 |
| Epi 6h | 425.047 | 124.773 | 1.48081 | 0.708022 |
| SA 6h | 282.96 | 43.0933 | 1.23087 | 0.104524 |
| Me-JA 6h | 141.682 | 53.5812 | 0.504811 | 0.250373 |
Source Transcript PGSC0003DMT400072450 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G28085.1 | +1 | 9e-19 | 77 | 41/81 (51%) | SAUR-like auxin-responsive protein family | chr2:11968182-11968556 REVERSE LENGTH=124 |
