Probe CUST_25492_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25492_PI426222305 | JHI_St_60k_v1 | DMT400022395 | CTCGAAATTGCCCTAATTATGCTGGATAACAAGAAGTTTGAAACTTTATCAAAAGAGGGG |
All Microarray Probes Designed to Gene DMG400008685
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_25697_PI426222305 | JHI_St_60k_v1 | DMT400022396 | GTCTTAGTGTAATGAGGGAGGAAAATAGTAAATTCTCAAAGTTTTAGCAAGTCCAGTGAA |
| CUST_25492_PI426222305 | JHI_St_60k_v1 | DMT400022395 | CTCGAAATTGCCCTAATTATGCTGGATAACAAGAAGTTTGAAACTTTATCAAAAGAGGGG |
Microarray Signals from CUST_25492_PI426222305
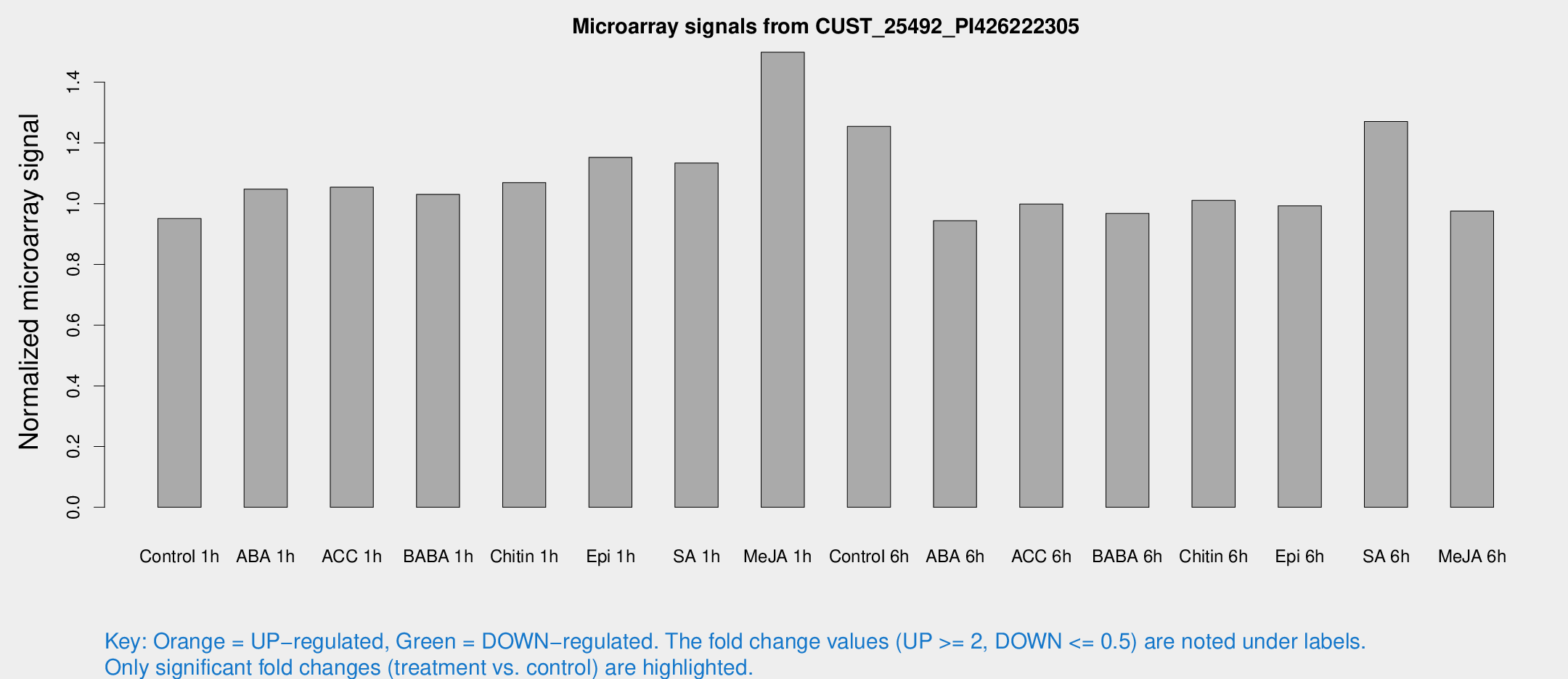
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.19083 | 3.0091 | 0.950663 | 0.545772 |
| ABA 1h | 5.021 | 2.9103 | 1.04797 | 0.596943 |
| ACC 1h | 6.0126 | 3.51545 | 1.05419 | 0.610451 |
| BABA 1h | 5.47482 | 3.17498 | 1.03067 | 0.596813 |
| Chitin 1h | 5.17677 | 3.01049 | 1.06931 | 0.604335 |
| Epi 1h | 5.4946 | 2.98293 | 1.15243 | 0.627423 |
| SA 1h | 6.68389 | 3.05288 | 1.13399 | 0.56326 |
| Me-JA 1h | 7.05641 | 3.12461 | 1.49859 | 0.732964 |
| Control 6h | 7.24815 | 3.11159 | 1.25436 | 0.590045 |
| ABA 6h | 5.52853 | 3.20458 | 0.944028 | 0.546649 |
| ACC 6h | 6.44679 | 3.82288 | 0.998458 | 0.578177 |
| BABA 6h | 5.97329 | 3.47044 | 0.967831 | 0.561082 |
| Chitin 6h | 5.92294 | 3.43261 | 1.0108 | 0.585614 |
| Epi 6h | 6.20362 | 3.62158 | 0.992765 | 0.575546 |
| SA 6h | 7.29857 | 3.29163 | 1.27048 | 0.630551 |
| Me-JA 6h | 5.35175 | 3.10191 | 0.975715 | 0.565014 |
Source Transcript PGSC0003DMT400022395 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G55940.1 | +1 | 1e-49 | 161 | 105/202 (52%) | Uncharacterised protein family (UPF0172) | chr5:22656256-22657894 REVERSE LENGTH=208 |
