Probe CUST_24320_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24320_PI426222305 | JHI_St_60k_v1 | DMT400074196 | CTACCTCCTGCTAACACAGGTTTCAAATAACTATTGTTACATGGTTATCATGACAAGTTC |
All Microarray Probes Designed to Gene DMG400028838
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24320_PI426222305 | JHI_St_60k_v1 | DMT400074196 | CTACCTCCTGCTAACACAGGTTTCAAATAACTATTGTTACATGGTTATCATGACAAGTTC |
Microarray Signals from CUST_24320_PI426222305
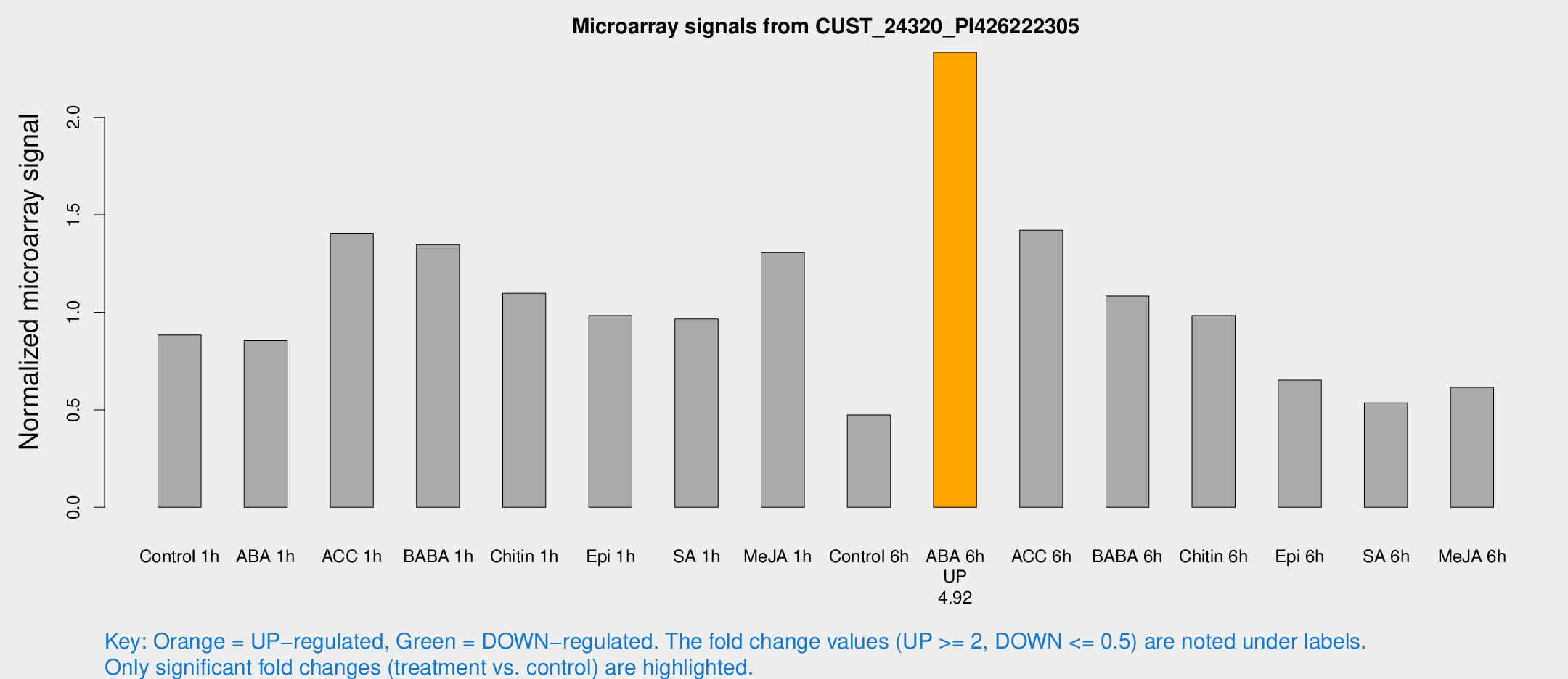
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 29.8257 | 4.8851 | 0.884044 | 0.163296 |
| ABA 1h | 27.5185 | 8.33318 | 0.855077 | 0.277747 |
| ACC 1h | 70.9585 | 30.3494 | 1.40528 | 1.42055 |
| BABA 1h | 42.85 | 4.52309 | 1.34714 | 0.238285 |
| Chitin 1h | 32.6599 | 4.14569 | 1.09759 | 0.140182 |
| Epi 1h | 28.8462 | 5.66668 | 0.983098 | 0.197028 |
| SA 1h | 32.6131 | 4.04529 | 0.965871 | 0.120507 |
| Me-JA 1h | 37.05 | 9.22134 | 1.3062 | 0.208462 |
| Control 6h | 17.8528 | 5.47982 | 0.47406 | 0.159179 |
| ABA 6h | 82.1648 | 9.26739 | 2.33357 | 0.170168 |
| ACC 6h | 53.6425 | 5.51408 | 1.42126 | 0.143188 |
| BABA 6h | 41.8706 | 10.0176 | 1.08383 | 0.229432 |
| Chitin 6h | 34.563 | 4.55903 | 0.982861 | 0.134494 |
| Epi 6h | 26.0618 | 6.78357 | 0.652161 | 0.246448 |
| SA 6h | 18.9404 | 5.79366 | 0.535665 | 0.144846 |
| Me-JA 6h | 25.6403 | 9.66857 | 0.615174 | 0.3558 |
Source Transcript PGSC0003DMT400074196 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
