Probe CUST_24037_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24037_PI426222305 | JHI_St_60k_v1 | DMT400027673 | GCTTCATGATGGTTGGCAAGAATTTATGGAACAATTAGTCTGTTGTCTATTGGTACTTTT |
All Microarray Probes Designed to Gene DMG400010668
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24037_PI426222305 | JHI_St_60k_v1 | DMT400027673 | GCTTCATGATGGTTGGCAAGAATTTATGGAACAATTAGTCTGTTGTCTATTGGTACTTTT |
| CUST_23944_PI426222305 | JHI_St_60k_v1 | DMT400027675 | GCTTCATGATGGTTGGCAAGAATTTATGGAACAATTAGTCTGTTGTCTATTGGTACTTTT |
Microarray Signals from CUST_24037_PI426222305
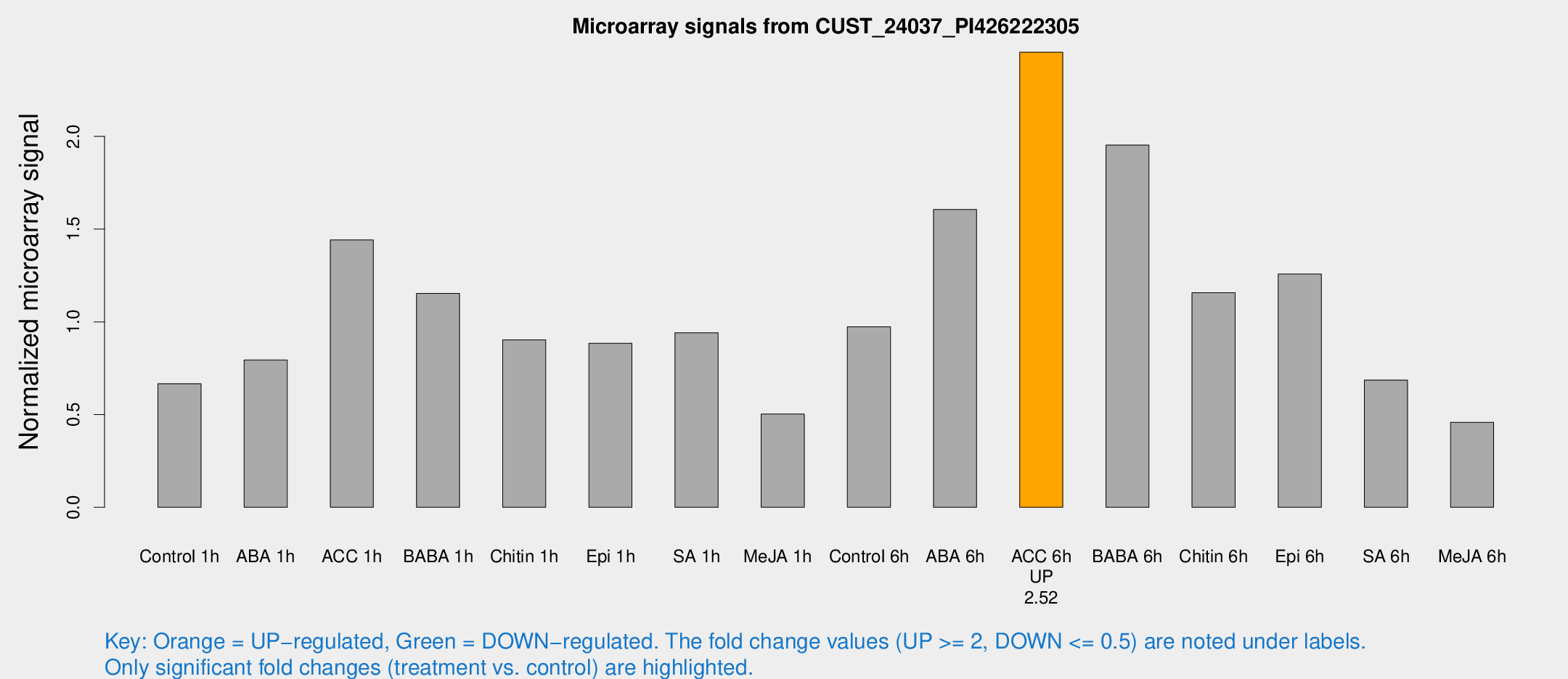
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 18.5655 | 5.22512 | 0.666329 | 0.26352 |
| ABA 1h | 19.0762 | 5.51856 | 0.793905 | 0.18201 |
| ACC 1h | 40.1558 | 10.7743 | 1.44204 | 0.343899 |
| BABA 1h | 29.8361 | 7.55349 | 1.15321 | 0.216025 |
| Chitin 1h | 25.1859 | 9.07565 | 0.902872 | 0.488941 |
| Epi 1h | 20.7112 | 5.97417 | 0.884365 | 0.264678 |
| SA 1h | 28.0151 | 11.1064 | 0.940922 | 0.32438 |
| Me-JA 1h | 11.5214 | 3.69136 | 0.503287 | 0.196098 |
| Control 6h | 25.0776 | 4.03339 | 0.973495 | 0.164165 |
| ABA 6h | 43.7962 | 6.96968 | 1.60548 | 0.187619 |
| ACC 6h | 70.8152 | 6.07561 | 2.45359 | 0.318439 |
| BABA 6h | 58.4709 | 16.1498 | 1.9528 | 0.455307 |
| Chitin 6h | 32.9533 | 8.55591 | 1.15704 | 0.367352 |
| Epi 6h | 37.3966 | 8.34737 | 1.25754 | 0.386232 |
| SA 6h | 17.14 | 3.83375 | 0.685949 | 0.158219 |
| Me-JA 6h | 12.7598 | 3.96143 | 0.458245 | 0.168364 |
Source Transcript PGSC0003DMT400027673 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G60142.1 | +2 | 2e-15 | 78 | 33/98 (34%) | AP2/B3-like transcriptional factor family protein | chr5:24216180-24217591 REVERSE LENGTH=346 |
