Probe CUST_24003_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24003_PI426222305 | JHI_St_60k_v1 | DMT400086558 | GTTGTTAACAATCAGATAAAAGCTGTAGACATGCTTTGCCAAGGTGACGAGATGGACATT |
All Microarray Probes Designed to Gene DMG400036129
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24003_PI426222305 | JHI_St_60k_v1 | DMT400086558 | GTTGTTAACAATCAGATAAAAGCTGTAGACATGCTTTGCCAAGGTGACGAGATGGACATT |
Microarray Signals from CUST_24003_PI426222305
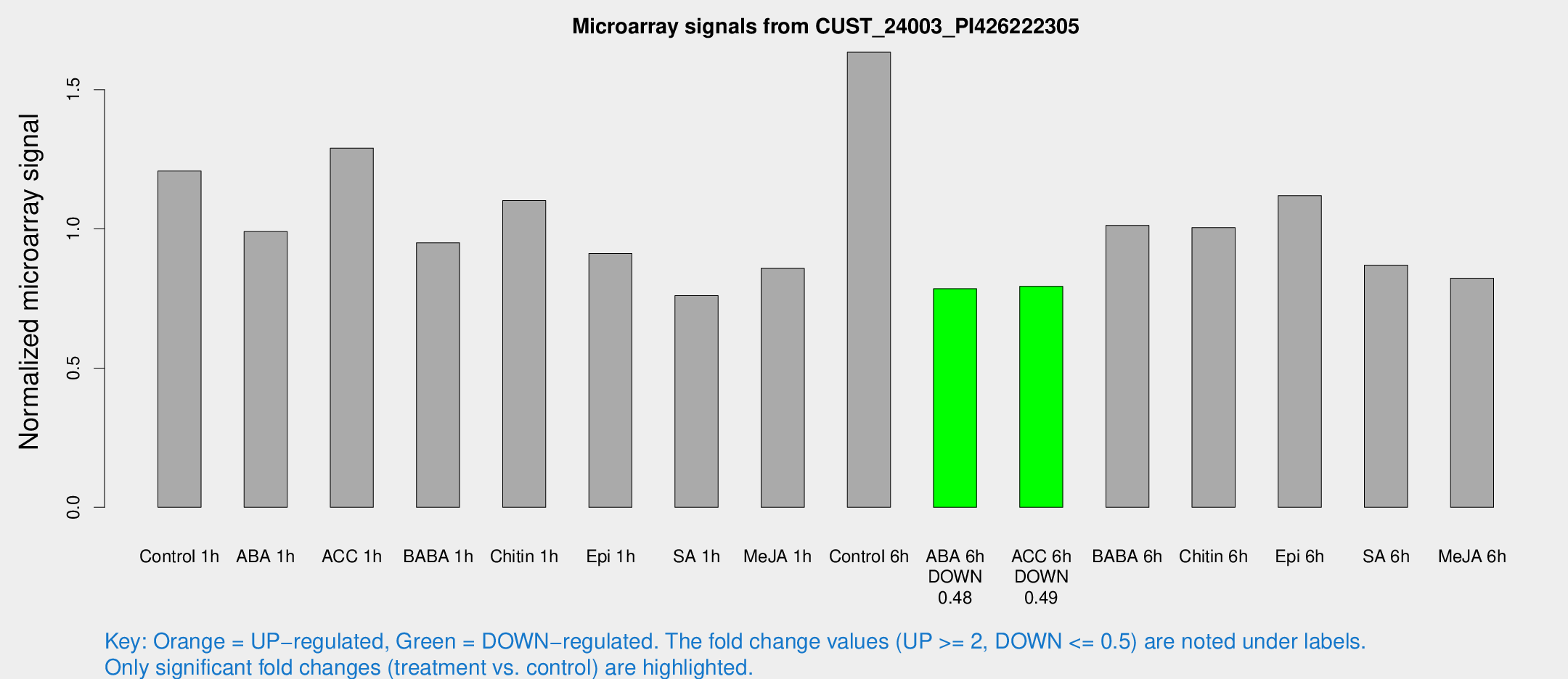
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 200.501 | 21.7115 | 1.20811 | 0.179941 |
| ABA 1h | 146.326 | 19.3261 | 0.990301 | 0.0653941 |
| ACC 1h | 217.964 | 13.1541 | 1.2903 | 0.0778761 |
| BABA 1h | 159.744 | 36.3983 | 0.950249 | 0.150257 |
| Chitin 1h | 164.724 | 18.8599 | 1.10176 | 0.110972 |
| Epi 1h | 130.779 | 13.4252 | 0.911717 | 0.105854 |
| SA 1h | 152.474 | 52.0883 | 0.760736 | 0.353306 |
| Me-JA 1h | 115.686 | 8.93103 | 0.85854 | 0.0730807 |
| Control 6h | 272.982 | 32.2474 | 1.63486 | 0.0967326 |
| ABA 6h | 138.285 | 12.8835 | 0.785397 | 0.0495348 |
| ACC 6h | 151.422 | 17.2202 | 0.793785 | 0.0523794 |
| BABA 6h | 195.026 | 39.039 | 1.01274 | 0.269742 |
| Chitin 6h | 176.347 | 12.1739 | 1.00493 | 0.0895269 |
| Epi 6h | 228.025 | 62.8758 | 1.11972 | 0.47924 |
| SA 6h | 161.267 | 49.1136 | 0.869963 | 0.254158 |
| Me-JA 6h | 147.414 | 44.7243 | 0.823321 | 0.186807 |
Source Transcript PGSC0003DMT400086558 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G26040.1 | +1 | 1e-65 | 219 | 141/430 (33%) | HXXXD-type acyl-transferase family protein | chr3:9519741-9521069 FORWARD LENGTH=442 |
