Probe CUST_23944_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23944_PI426222305 | JHI_St_60k_v1 | DMT400027675 | GCTTCATGATGGTTGGCAAGAATTTATGGAACAATTAGTCTGTTGTCTATTGGTACTTTT |
All Microarray Probes Designed to Gene DMG400010668
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_24037_PI426222305 | JHI_St_60k_v1 | DMT400027673 | GCTTCATGATGGTTGGCAAGAATTTATGGAACAATTAGTCTGTTGTCTATTGGTACTTTT |
| CUST_23944_PI426222305 | JHI_St_60k_v1 | DMT400027675 | GCTTCATGATGGTTGGCAAGAATTTATGGAACAATTAGTCTGTTGTCTATTGGTACTTTT |
Microarray Signals from CUST_23944_PI426222305
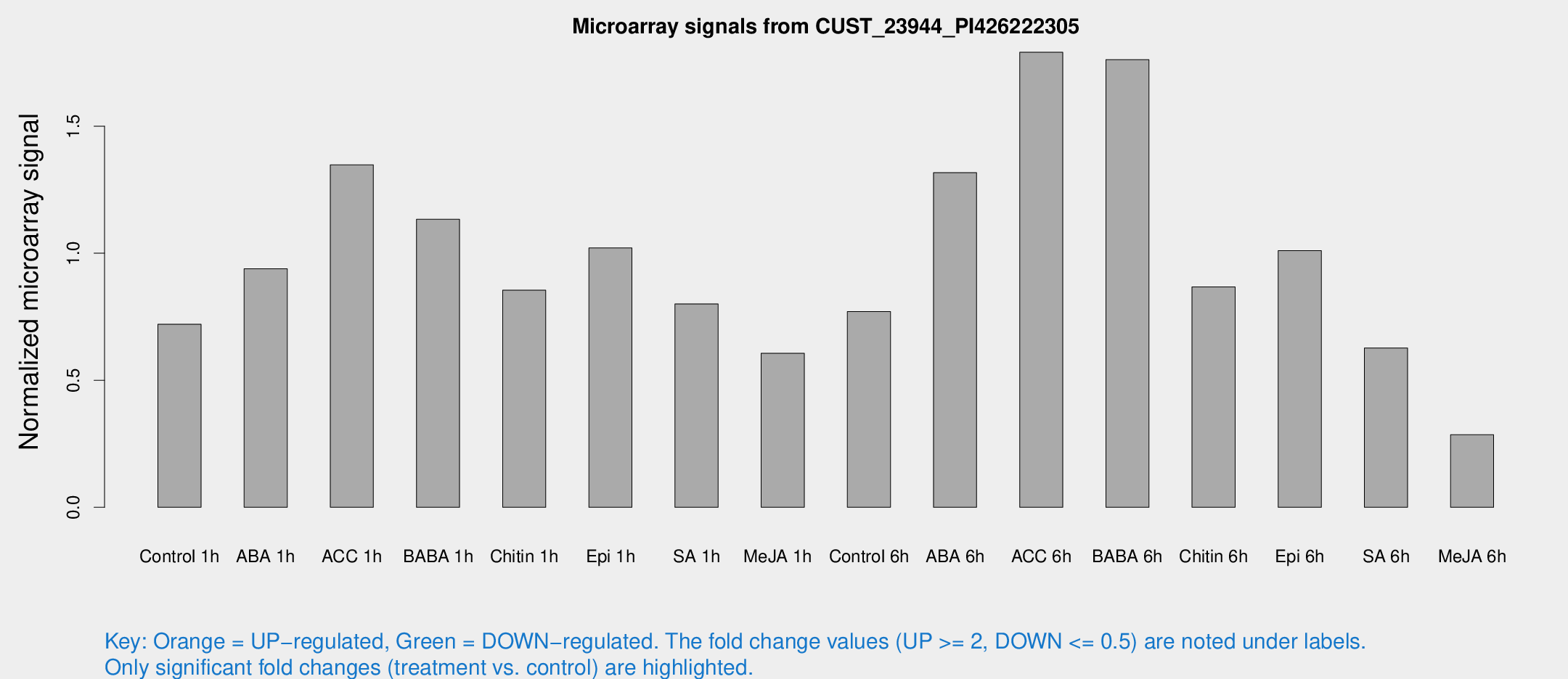
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 21.1777 | 4.93693 | 0.720741 | 0.257724 |
| ABA 1h | 24.4146 | 6.37394 | 0.939173 | 0.237864 |
| ACC 1h | 38.4148 | 4.49875 | 1.34793 | 0.167703 |
| BABA 1h | 32.3631 | 8.73877 | 1.13358 | 0.226131 |
| Chitin 1h | 22.4535 | 5.85717 | 0.854422 | 0.149641 |
| Epi 1h | 24.6638 | 3.5229 | 1.02071 | 0.152343 |
| SA 1h | 25.0418 | 6.99649 | 0.800638 | 0.241949 |
| Me-JA 1h | 16.1032 | 5.3239 | 0.606204 | 0.257546 |
| Control 6h | 23.5874 | 6.57738 | 0.770698 | 0.204638 |
| ABA 6h | 42.0144 | 11.3994 | 1.31702 | 0.357587 |
| ACC 6h | 56.4512 | 5.2817 | 1.79122 | 0.222848 |
| BABA 6h | 54.2936 | 4.86252 | 1.76204 | 0.157481 |
| Chitin 6h | 31.0188 | 10.8737 | 0.86724 | 0.576638 |
| Epi 6h | 31.3781 | 4.3409 | 1.01027 | 0.142381 |
| SA 6h | 18.6547 | 5.0466 | 0.627004 | 0.25542 |
| Me-JA 6h | 8.71792 | 3.39416 | 0.285608 | 0.135862 |
Source Transcript PGSC0003DMT400027675 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G60140.1 | +1 | 2e-09 | 57 | 32/98 (33%) | AP2/B3-like transcriptional factor family protein | chr5:24214077-24215351 REVERSE LENGTH=328 |
