Probe CUST_23794_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23794_PI426222305 | JHI_St_60k_v1 | DMT400021142 | GCTGTTTAATGCCGTGCAATGCTTTCTGTTACATTTTGTTTTTCTTTAAACAGAAATGGC |
All Microarray Probes Designed to Gene DMG400008187
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23794_PI426222305 | JHI_St_60k_v1 | DMT400021142 | GCTGTTTAATGCCGTGCAATGCTTTCTGTTACATTTTGTTTTTCTTTAAACAGAAATGGC |
Microarray Signals from CUST_23794_PI426222305
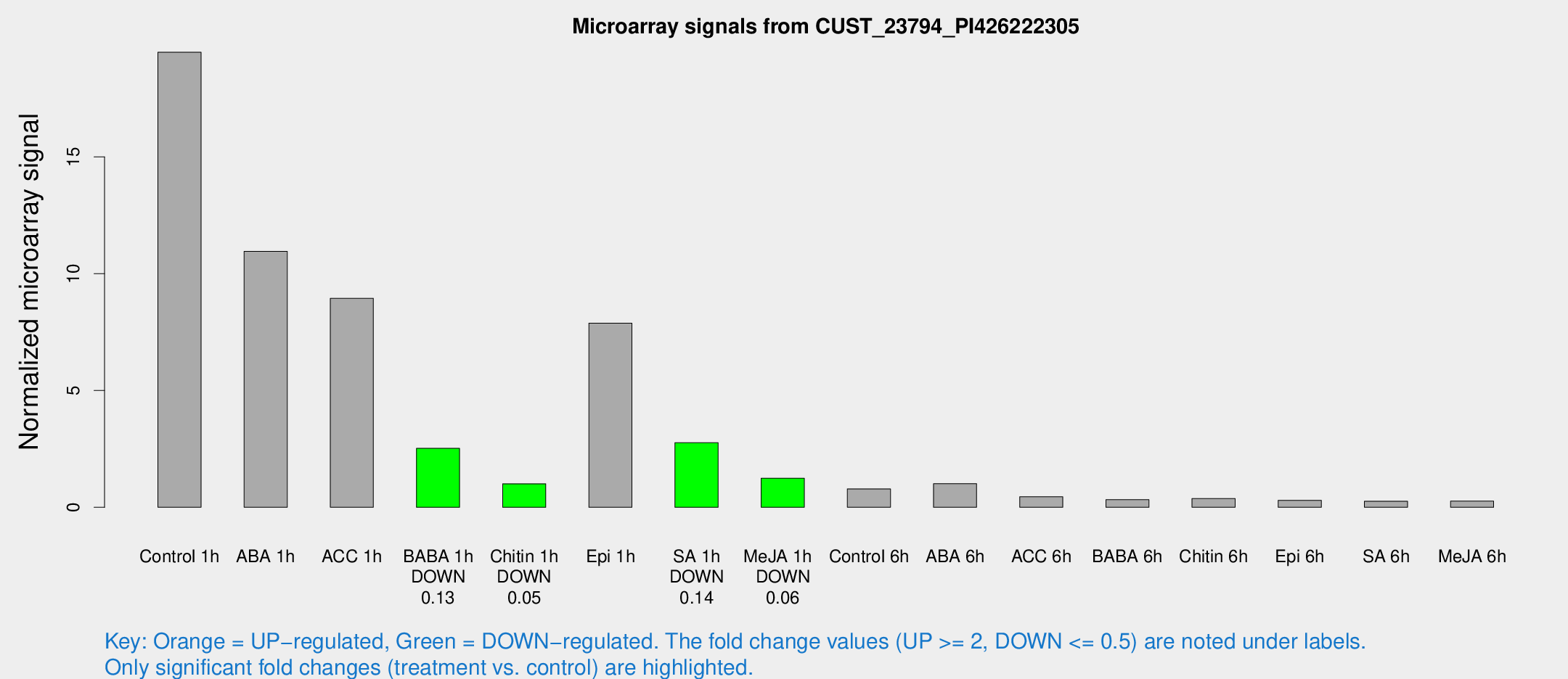
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3775.05 | 720.017 | 19.4828 | 3.73405 |
| ABA 1h | 3677.44 | 2853.68 | 10.9578 | 17.7632 |
| ACC 1h | 2776.03 | 1669.79 | 8.94667 | 8.94349 |
| BABA 1h | 531.904 | 200.581 | 2.52345 | 0.847372 |
| Chitin 1h | 190.161 | 67.3805 | 1.00774 | 0.277243 |
| Epi 1h | 1654 | 820.612 | 7.8848 | 5.2068 |
| SA 1h | 669.53 | 283.69 | 2.76341 | 1.56235 |
| Me-JA 1h | 191.416 | 11.5351 | 1.24733 | 0.136895 |
| Control 6h | 158.706 | 41.443 | 0.790128 | 0.163825 |
| ABA 6h | 203.692 | 19.8097 | 1.01296 | 0.155869 |
| ACC 6h | 101.515 | 19.785 | 0.452391 | 0.169942 |
| BABA 6h | 69.9396 | 10.2649 | 0.325796 | 0.0645948 |
| Chitin 6h | 75.2221 | 7.03359 | 0.373716 | 0.0305533 |
| Epi 6h | 63.5342 | 5.23282 | 0.299054 | 0.0245297 |
| SA 6h | 59.6252 | 21.4601 | 0.259147 | 0.117881 |
| Me-JA 6h | 64.1526 | 26.5151 | 0.264867 | 0.192424 |
Source Transcript PGSC0003DMT400021142 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G12020.1 | +2 | 2e-45 | 153 | 96/149 (64%) | 17.6 kDa class II heat shock protein | chr5:3882409-3882876 REVERSE LENGTH=155 |
