Probe CUST_23763_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23763_PI426222305 | JHI_St_60k_v1 | DMT400021236 | TCCTAAGCTTGTGGAAGTCGGTGTCATCACTCATGTGGTATAGTAAGTAAAATTTCCATG |
All Microarray Probes Designed to Gene DMG400008224
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23763_PI426222305 | JHI_St_60k_v1 | DMT400021236 | TCCTAAGCTTGTGGAAGTCGGTGTCATCACTCATGTGGTATAGTAAGTAAAATTTCCATG |
Microarray Signals from CUST_23763_PI426222305
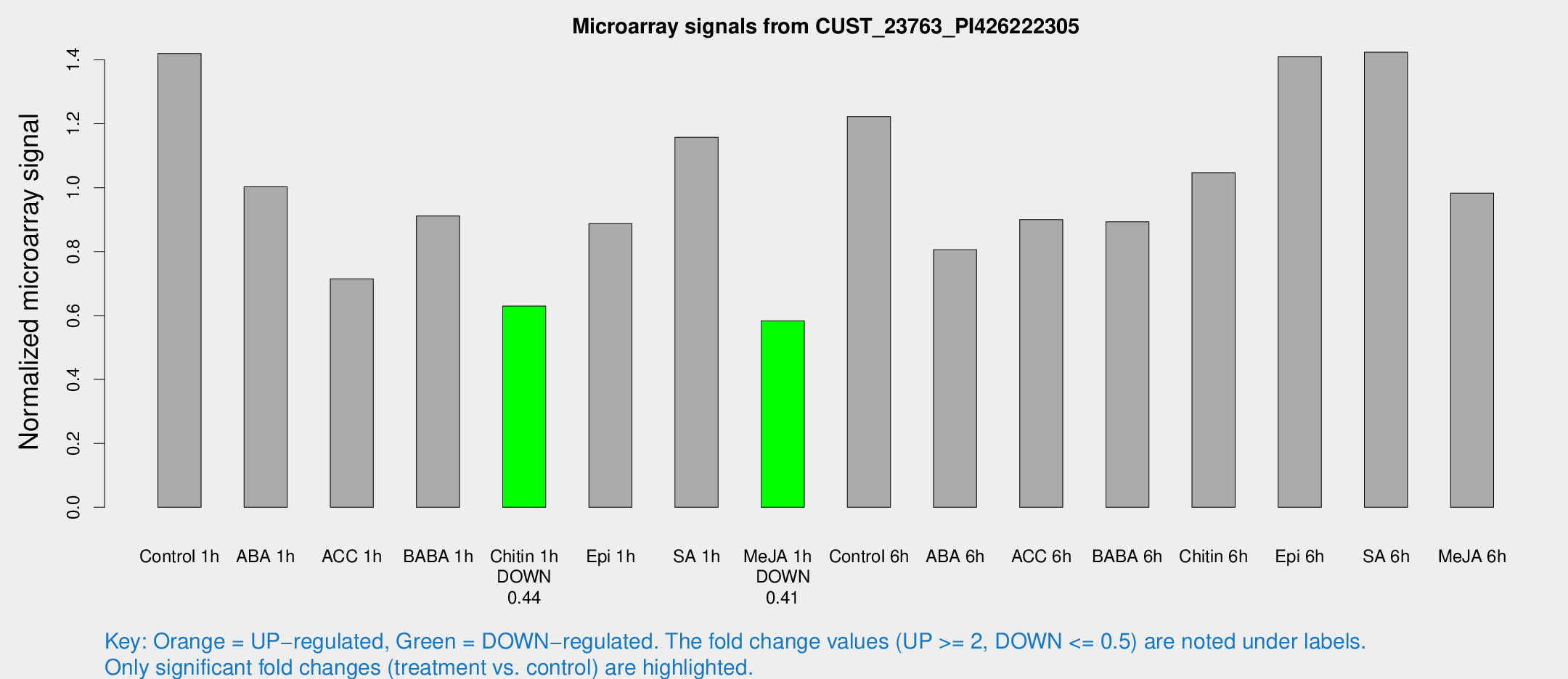
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3812.09 | 404.625 | 1.41978 | 0.0819829 |
| ABA 1h | 2391.64 | 283.478 | 1.0026 | 0.0897804 |
| ACC 1h | 2643.69 | 1102.9 | 0.714705 | 0.506038 |
| BABA 1h | 2441.48 | 490.071 | 0.91159 | 0.110835 |
| Chitin 1h | 1557.12 | 301.041 | 0.629618 | 0.0873406 |
| Epi 1h | 2080.77 | 275.456 | 0.887636 | 0.115228 |
| SA 1h | 3226.77 | 472.688 | 1.15766 | 0.149736 |
| Me-JA 1h | 1277.69 | 123.181 | 0.583527 | 0.0337351 |
| Control 6h | 3455.15 | 883.873 | 1.22244 | 0.260708 |
| ABA 6h | 2278.45 | 131.797 | 0.805818 | 0.0465449 |
| ACC 6h | 2751.72 | 168.839 | 0.89984 | 0.134686 |
| BABA 6h | 2660.2 | 153.887 | 0.893374 | 0.0515981 |
| Chitin 6h | 2962.75 | 171.298 | 1.04683 | 0.0604572 |
| Epi 6h | 4398.65 | 883.418 | 1.4103 | 0.180727 |
| SA 6h | 3778.37 | 391.154 | 1.42368 | 0.129218 |
| Me-JA 6h | 2639.38 | 325.854 | 0.983073 | 0.11128 |
Source Transcript PGSC0003DMT400021236 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G33040.1 | +3 | 1e-40 | 139 | 74/125 (59%) | Thioredoxin superfamily protein | chr4:15940779-15941213 REVERSE LENGTH=144 |
