Probe CUST_23011_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_23011_PI426222305 | JHI_St_60k_v1 | DMT400061003 | CAACCTGGGGTATTGAATAAATTCAAGGATATTCATGCATGATGCATTTGAAGTCAAATG |
All Microarray Probes Designed to Gene DMG400023732
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22915_PI426222305 | JHI_St_60k_v1 | DMT400061002 | GTGGCAGAGGGTGGAATTAAACGTATAGAATTGGATTCATTCATATCTCATATAACAAGA |
| CUST_23011_PI426222305 | JHI_St_60k_v1 | DMT400061003 | CAACCTGGGGTATTGAATAAATTCAAGGATATTCATGCATGATGCATTTGAAGTCAAATG |
Microarray Signals from CUST_23011_PI426222305
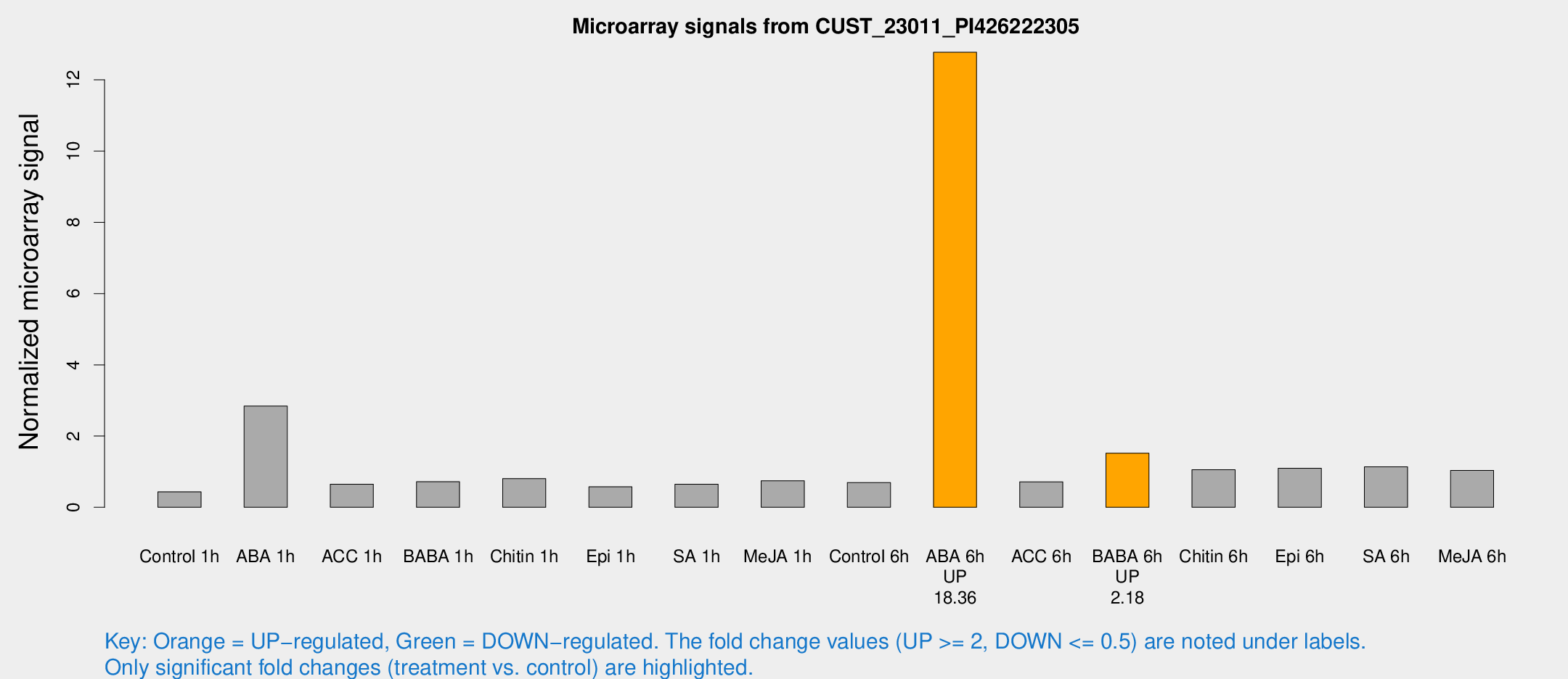
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 11.2183 | 3.66265 | 0.435258 | 0.147934 |
| ABA 1h | 71.7858 | 22.003 | 2.84366 | 0.880147 |
| ACC 1h | 18.2086 | 4.7894 | 0.648136 | 0.180099 |
| BABA 1h | 20.5104 | 6.60231 | 0.717825 | 0.250552 |
| Chitin 1h | 19.6687 | 4.62896 | 0.806636 | 0.212411 |
| Epi 1h | 15.9359 | 7.75835 | 0.575189 | 0.337378 |
| SA 1h | 18.2907 | 4.63693 | 0.646433 | 0.171532 |
| Me-JA 1h | 17.3421 | 4.96118 | 0.745994 | 0.378176 |
| Control 6h | 18.3712 | 4.05794 | 0.695984 | 0.170783 |
| ABA 6h | 356.457 | 59.49 | 12.7756 | 1.71626 |
| ACC 6h | 24.0563 | 8.94525 | 0.715182 | 0.189226 |
| BABA 6h | 44.2785 | 6.97432 | 1.52013 | 0.223425 |
| Chitin 6h | 30.4861 | 6.97174 | 1.05388 | 0.339044 |
| Epi 6h | 32.8461 | 7.06439 | 1.09818 | 0.23738 |
| SA 6h | 31.1407 | 8.65746 | 1.13787 | 0.225851 |
| Me-JA 6h | 31.745 | 10.8917 | 1.03708 | 0.481336 |
Source Transcript PGSC0003DMT400061003 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G34135.1 | +1 | 1e-50 | 183 | 141/496 (28%) | UDP-glucosyltransferase 73B2 | chr4:16345476-16347016 REVERSE LENGTH=483 |
