Probe CUST_22808_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22808_PI426222305 | JHI_St_60k_v1 | DMT400078314 | TTGTGATAGAAGTTAGGAAAAGAATGTTTTCTTTTTTAGGGGATATGTGCTCTCTTGAGG |
All Microarray Probes Designed to Gene DMG400030486
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22808_PI426222305 | JHI_St_60k_v1 | DMT400078314 | TTGTGATAGAAGTTAGGAAAAGAATGTTTTCTTTTTTAGGGGATATGTGCTCTCTTGAGG |
Microarray Signals from CUST_22808_PI426222305
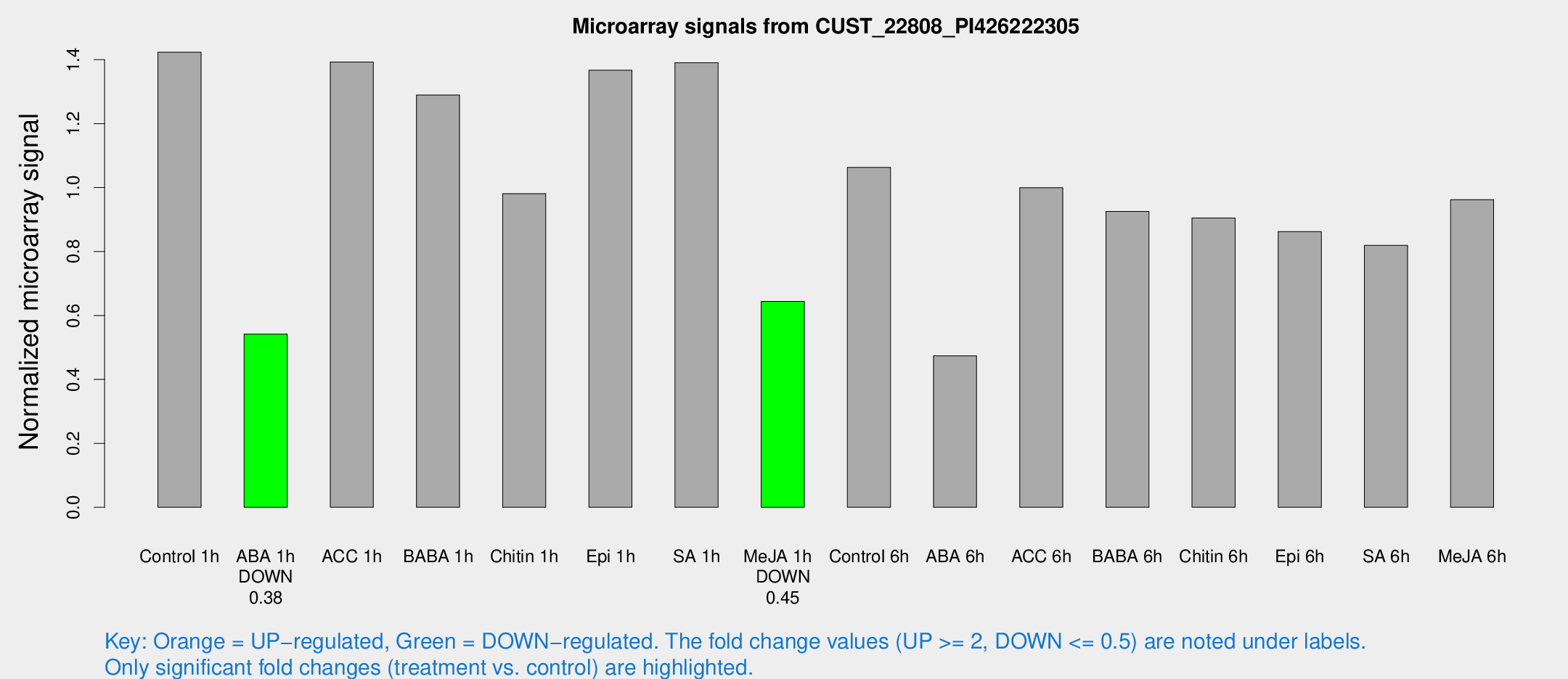
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 970.58 | 121.78 | 1.42333 | 0.0918538 |
| ABA 1h | 323.74 | 23.7217 | 0.541876 | 0.0591721 |
| ACC 1h | 1006.24 | 198.219 | 1.39279 | 0.202178 |
| BABA 1h | 852.635 | 115.788 | 1.28947 | 0.0746275 |
| Chitin 1h | 593.94 | 34.4962 | 0.980899 | 0.0568479 |
| Epi 1h | 802.955 | 81.6559 | 1.36719 | 0.146604 |
| SA 1h | 969.32 | 95.9357 | 1.39062 | 0.0804155 |
| Me-JA 1h | 355.884 | 31.0687 | 0.644084 | 0.037616 |
| Control 6h | 730.198 | 97.4202 | 1.063 | 0.061564 |
| ABA 6h | 346.603 | 55.8713 | 0.473676 | 0.0500481 |
| ACC 6h | 772.711 | 44.8943 | 0.99978 | 0.0867987 |
| BABA 6h | 701.991 | 64.4107 | 0.925089 | 0.0536155 |
| Chitin 6h | 655.825 | 73.4841 | 0.904743 | 0.0795071 |
| Epi 6h | 654.23 | 37.9937 | 0.86252 | 0.0533683 |
| SA 6h | 590.005 | 146.951 | 0.819493 | 0.151201 |
| Me-JA 6h | 647.136 | 45.0247 | 0.962317 | 0.0557563 |
Source Transcript PGSC0003DMT400078314 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G07300.1 | +2 | 2e-06 | 45 | 31/68 (46%) | josephin protein-related | chr1:2245914-2246300 REVERSE LENGTH=128 |
