Probe CUST_22553_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22553_PI426222305 | JHI_St_60k_v1 | DMT400078081 | ATCAAAGTGTATCAATTTGAAGGCTTCATCTTCTTCTTATTCTGCATTCTCCGATCCCTT |
All Microarray Probes Designed to Gene DMG400030370
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22587_PI426222305 | JHI_St_60k_v1 | DMT400078083 | ATCAAAGTGTATCAATTTGAAGGCTTCATCTTCTTCTTATTCTGCATTCTCCGATCCCTT |
| CUST_22553_PI426222305 | JHI_St_60k_v1 | DMT400078081 | ATCAAAGTGTATCAATTTGAAGGCTTCATCTTCTTCTTATTCTGCATTCTCCGATCCCTT |
Microarray Signals from CUST_22553_PI426222305
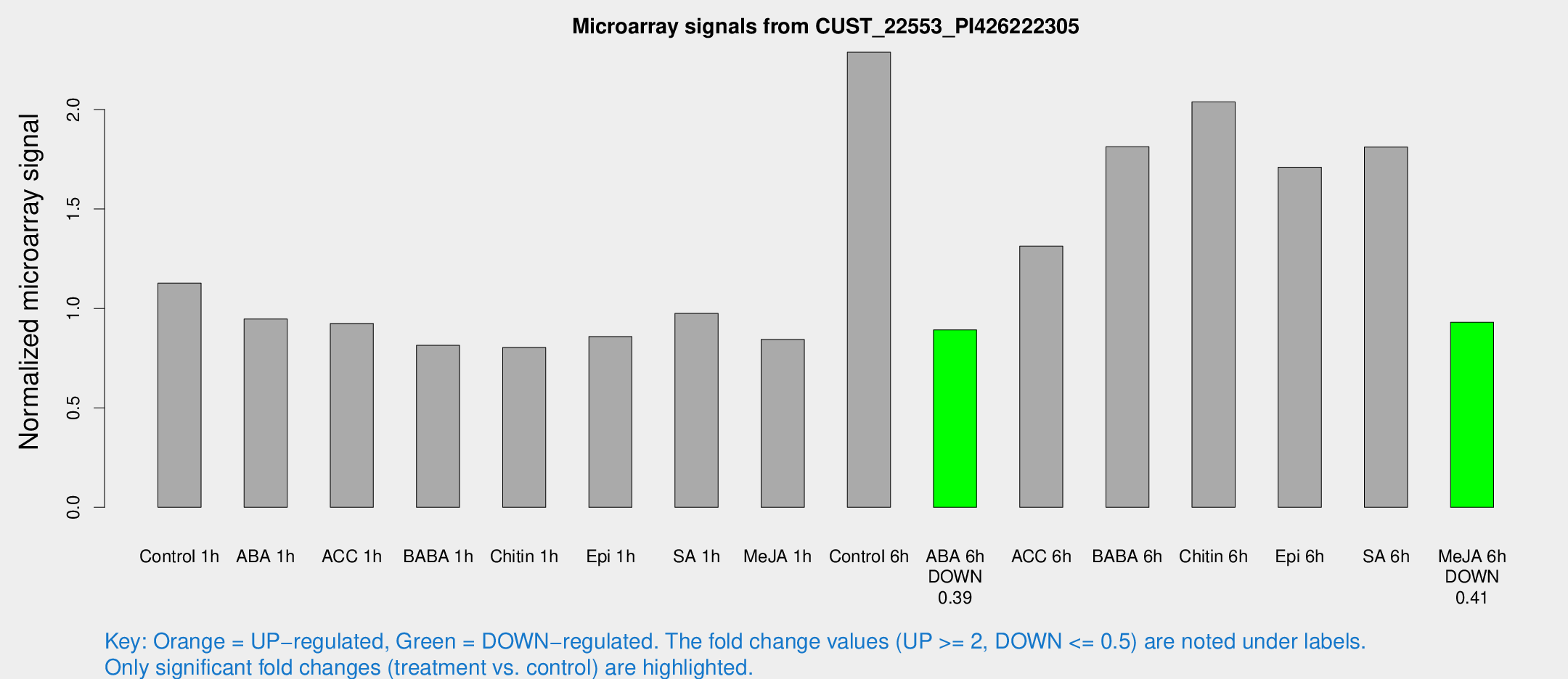
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 784.167 | 102.146 | 1.12731 | 0.0977614 |
| ABA 1h | 575.093 | 33.4988 | 0.946548 | 0.054968 |
| ACC 1h | 672.765 | 116.292 | 0.924071 | 0.106798 |
| BABA 1h | 558.545 | 101.901 | 0.814481 | 0.0814684 |
| Chitin 1h | 498.218 | 43.2453 | 0.803462 | 0.0467466 |
| Epi 1h | 514.872 | 56.7545 | 0.858262 | 0.0772127 |
| SA 1h | 690.097 | 58.0597 | 0.974532 | 0.0564866 |
| Me-JA 1h | 476.279 | 47.0243 | 0.84339 | 0.0491256 |
| Control 6h | 1673.6 | 398.832 | 2.28791 | 0.426622 |
| ABA 6h | 654.428 | 60.2277 | 0.891835 | 0.0517678 |
| ACC 6h | 1108.67 | 274.318 | 1.31324 | 0.25037 |
| BABA 6h | 1436.3 | 274.255 | 1.81274 | 0.327047 |
| Chitin 6h | 1502.42 | 165.98 | 2.03782 | 0.177935 |
| Epi 6h | 1333.88 | 137.763 | 1.7096 | 0.268806 |
| SA 6h | 1227.45 | 71.0307 | 1.81141 | 0.211585 |
| Me-JA 6h | 704.619 | 217.083 | 0.930459 | 0.241123 |
Source Transcript PGSC0003DMT400078081 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G07110.1 | +3 | 0.0 | 1101 | 573/752 (76%) | fructose-2,6-bisphosphatase | chr1:2178363-2183980 REVERSE LENGTH=744 |
