Probe CUST_22546_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22546_PI426222305 | JHI_St_60k_v1 | DMT400078290 | CACATTTTGAAGCATTCCACCATTGTGATGAAACCAAGATCTTTCTAAACTTTGTAATGC |
All Microarray Probes Designed to Gene DMG402030469
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22546_PI426222305 | JHI_St_60k_v1 | DMT400078290 | CACATTTTGAAGCATTCCACCATTGTGATGAAACCAAGATCTTTCTAAACTTTGTAATGC |
Microarray Signals from CUST_22546_PI426222305
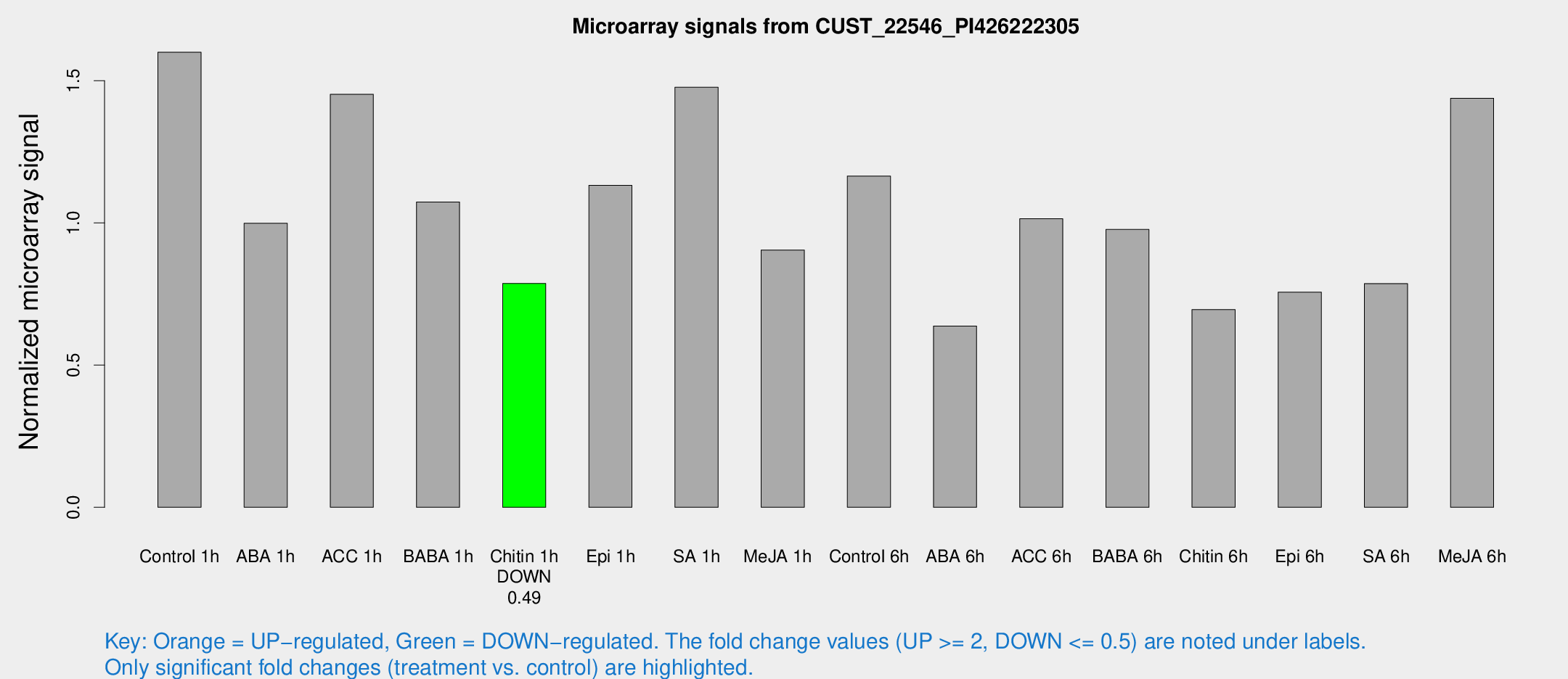
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14934.7 | 1928.58 | 1.60031 | 0.0988942 |
| ABA 1h | 8139.17 | 501.68 | 0.99866 | 0.0576588 |
| ACC 1h | 15280.9 | 4295 | 1.45226 | 0.428825 |
| BABA 1h | 9660.35 | 1183.24 | 1.07378 | 0.061996 |
| Chitin 1h | 6557.67 | 613.464 | 0.787103 | 0.0522985 |
| Epi 1h | 9008.59 | 520.882 | 1.13222 | 0.0653699 |
| SA 1h | 14826.2 | 3875.97 | 1.47697 | 0.370493 |
| Me-JA 1h | 6831.48 | 590.967 | 0.904843 | 0.0522428 |
| Control 6h | 11116.5 | 1962.57 | 1.16503 | 0.119008 |
| ABA 6h | 6300.17 | 702.491 | 0.637583 | 0.0586464 |
| ACC 6h | 11112.3 | 2038.22 | 1.01483 | 0.234674 |
| BABA 6h | 10178.6 | 1214.8 | 0.976944 | 0.102398 |
| Chitin 6h | 6859.78 | 682.528 | 0.695073 | 0.0839506 |
| Epi 6h | 8037.59 | 1217.87 | 0.756477 | 0.181641 |
| SA 6h | 7165.6 | 415.011 | 0.786616 | 0.052337 |
| Me-JA 6h | 13216.8 | 947.657 | 1.43833 | 0.133037 |
Source Transcript PGSC0003DMT400078290 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G30490.1 | +2 | 0.0 | 824 | 437/504 (87%) | cinnamate-4-hydroxylase | chr2:12993861-12995683 REVERSE LENGTH=505 |
