Probe CUST_22324_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22324_PI426222305 | JHI_St_60k_v1 | DMT400039340 | CATCTTCATTATTCATGTTCATAAGCCAACCATTGGCTGTTGGTTACAATTTGGTTATGC |
All Microarray Probes Designed to Gene DMG400015214
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22324_PI426222305 | JHI_St_60k_v1 | DMT400039340 | CATCTTCATTATTCATGTTCATAAGCCAACCATTGGCTGTTGGTTACAATTTGGTTATGC |
Microarray Signals from CUST_22324_PI426222305
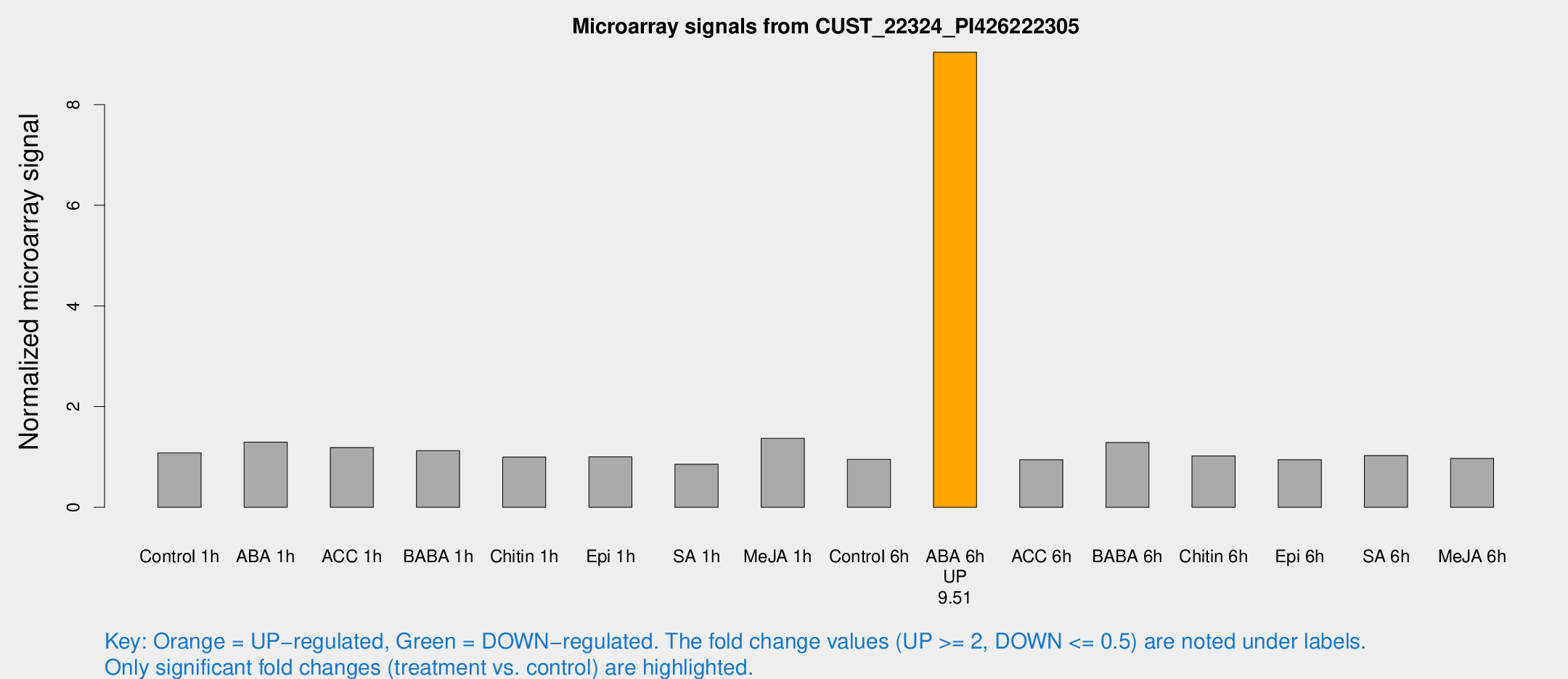
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.9035 | 3.60677 | 1.08194 | 0.538921 |
| ABA 1h | 8.77258 | 3.50874 | 1.29502 | 0.62038 |
| ACC 1h | 8.89039 | 3.96096 | 1.18779 | 0.583713 |
| BABA 1h | 7.83653 | 3.85365 | 1.12517 | 0.581726 |
| Chitin 1h | 6.28836 | 3.6638 | 0.998531 | 0.578307 |
| Epi 1h | 6.05394 | 3.51558 | 1.00182 | 0.580127 |
| SA 1h | 6.13267 | 3.5619 | 0.855916 | 0.496229 |
| Me-JA 1h | 8.4222 | 3.76556 | 1.36954 | 0.681558 |
| Control 6h | 6.64655 | 3.8548 | 0.951161 | 0.550813 |
| ABA 6h | 72.7485 | 20.9846 | 9.04259 | 2.19695 |
| ACC 6h | 7.70464 | 4.55379 | 0.946236 | 0.548043 |
| BABA 6h | 11.7638 | 4.9366 | 1.28687 | 0.612141 |
| Chitin 6h | 7.57315 | 4.39393 | 1.02047 | 0.591241 |
| Epi 6h | 7.52394 | 4.41571 | 0.946327 | 0.547976 |
| SA 6h | 7.08253 | 4.10291 | 1.02833 | 0.595573 |
| Me-JA 6h | 6.74113 | 3.91393 | 0.970601 | 0.562479 |
Source Transcript PGSC0003DMT400039340 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G54540.1 | +2 | 6e-66 | 211 | 125/258 (48%) | Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family | chr1:20367702-20368421 REVERSE LENGTH=239 |
