Probe CUST_22084_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22084_PI426222305 | JHI_St_60k_v1 | DMT400077708 | ATACCTTGTCTTTATGTTTTCATGTGGTCATGTTTGAGGAAATAATCTCTTGTGGTCACT |
All Microarray Probes Designed to Gene DMG400030231
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_22084_PI426222305 | JHI_St_60k_v1 | DMT400077708 | ATACCTTGTCTTTATGTTTTCATGTGGTCATGTTTGAGGAAATAATCTCTTGTGGTCACT |
Microarray Signals from CUST_22084_PI426222305
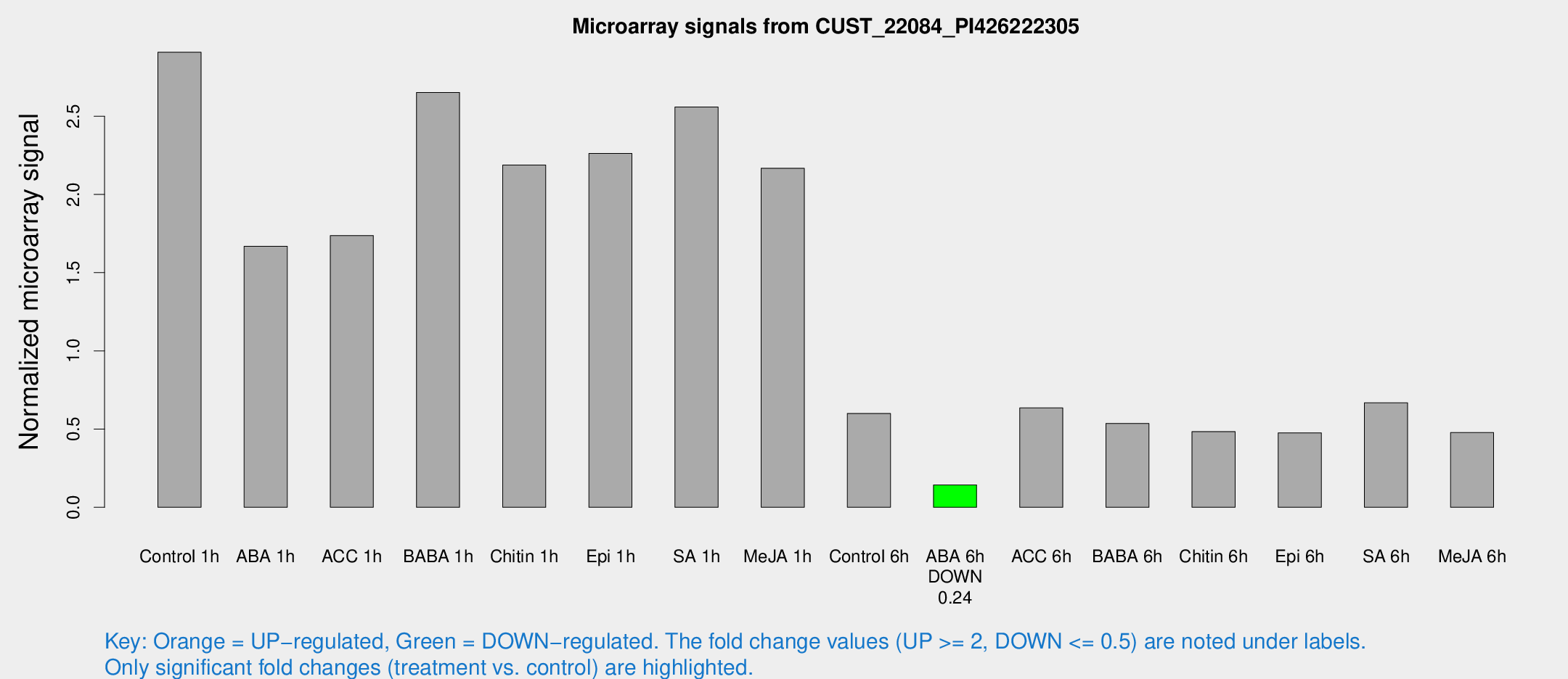
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3472.35 | 200.999 | 2.90909 | 0.167984 |
| ABA 1h | 1778.89 | 193.578 | 1.66888 | 0.152905 |
| ACC 1h | 2249.12 | 489.568 | 1.73738 | 0.300662 |
| BABA 1h | 3149.17 | 539.45 | 2.65204 | 0.227752 |
| Chitin 1h | 2347.54 | 135.868 | 2.18774 | 0.126355 |
| Epi 1h | 2386.25 | 344.783 | 2.26166 | 0.333721 |
| SA 1h | 3149.84 | 242.664 | 2.55939 | 0.157669 |
| Me-JA 1h | 2145.84 | 296.73 | 2.16764 | 0.125207 |
| Control 6h | 745.099 | 139.621 | 0.600305 | 0.0747752 |
| ABA 6h | 181.5 | 11.2527 | 0.143042 | 0.0088481 |
| ACC 6h | 873.267 | 71.8423 | 0.635156 | 0.0373488 |
| BABA 6h | 720.507 | 65.2204 | 0.536201 | 0.0399789 |
| Chitin 6h | 616.004 | 41.3114 | 0.483878 | 0.0281593 |
| Epi 6h | 644.36 | 57.2004 | 0.476182 | 0.0653165 |
| SA 6h | 802.063 | 109.05 | 0.667679 | 0.0387141 |
| Me-JA 6h | 571.135 | 53.1016 | 0.477438 | 0.0364448 |
Source Transcript PGSC0003DMT400077708 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00880.1 | +2 | 1e-26 | 102 | 49/86 (57%) | SAUR-like auxin-responsive protein family | chr4:366692-367060 REVERSE LENGTH=122 |
