Probe CUST_21518_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21518_PI426222305 | JHI_St_60k_v1 | DMT400093652 | TATATGATGAATCCGGATGGTAATGGAGGACCTGAACTCAGTATTTTTCTTCTTAGAATA |
All Microarray Probes Designed to Gene DMG400043223
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21518_PI426222305 | JHI_St_60k_v1 | DMT400093652 | TATATGATGAATCCGGATGGTAATGGAGGACCTGAACTCAGTATTTTTCTTCTTAGAATA |
Microarray Signals from CUST_21518_PI426222305
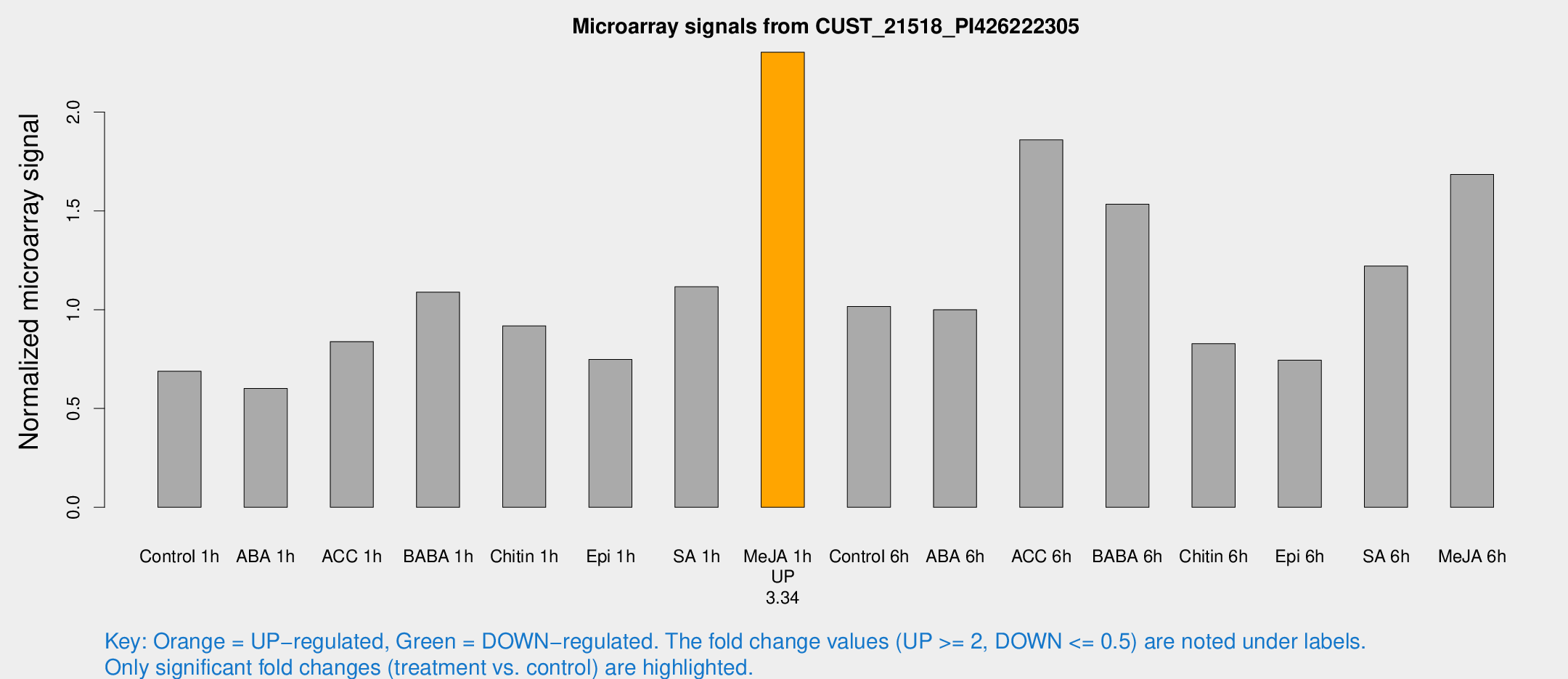
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 153.784 | 10.3151 | 0.688717 | 0.0423391 |
| ABA 1h | 120.036 | 15.0145 | 0.601037 | 0.038437 |
| ACC 1h | 206.057 | 50.3312 | 0.838608 | 0.182761 |
| BABA 1h | 234.374 | 16.5548 | 1.08888 | 0.100973 |
| Chitin 1h | 187.904 | 28.2766 | 0.9183 | 0.117857 |
| Epi 1h | 147.49 | 23.7527 | 0.74852 | 0.0955181 |
| SA 1h | 280.751 | 91.651 | 1.11619 | 0.378325 |
| Me-JA 1h | 421.298 | 38.7795 | 2.3029 | 0.236512 |
| Control 6h | 235.169 | 43.4355 | 1.01595 | 0.122882 |
| ABA 6h | 238.733 | 23.5846 | 0.999777 | 0.117275 |
| ACC 6h | 485.059 | 72.1061 | 1.85941 | 0.40798 |
| BABA 6h | 395.729 | 77.8435 | 1.53362 | 0.266346 |
| Chitin 6h | 198.499 | 23.3755 | 0.828134 | 0.102763 |
| Epi 6h | 187.023 | 11.5314 | 0.744678 | 0.0457704 |
| SA 6h | 269.539 | 17.5663 | 1.22112 | 0.137926 |
| Me-JA 6h | 377.012 | 39.5932 | 1.68461 | 0.292785 |
Source Transcript PGSC0003DMT400093652 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G41660.1 | +1 | 3e-78 | 243 | 131/211 (62%) | Protein of unknown function, DUF617 | chr2:17367945-17368838 FORWARD LENGTH=297 |
