Probe CUST_21503_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21503_PI426222305 | JHI_St_60k_v1 | DMT400070176 | TTTGGTCTTCCAGACAACAGAAGAATGGTTGAACAGCACTTTGGGGTTGCACCCAATTCA |
All Microarray Probes Designed to Gene DMG400027276
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21503_PI426222305 | JHI_St_60k_v1 | DMT400070176 | TTTGGTCTTCCAGACAACAGAAGAATGGTTGAACAGCACTTTGGGGTTGCACCCAATTCA |
Microarray Signals from CUST_21503_PI426222305
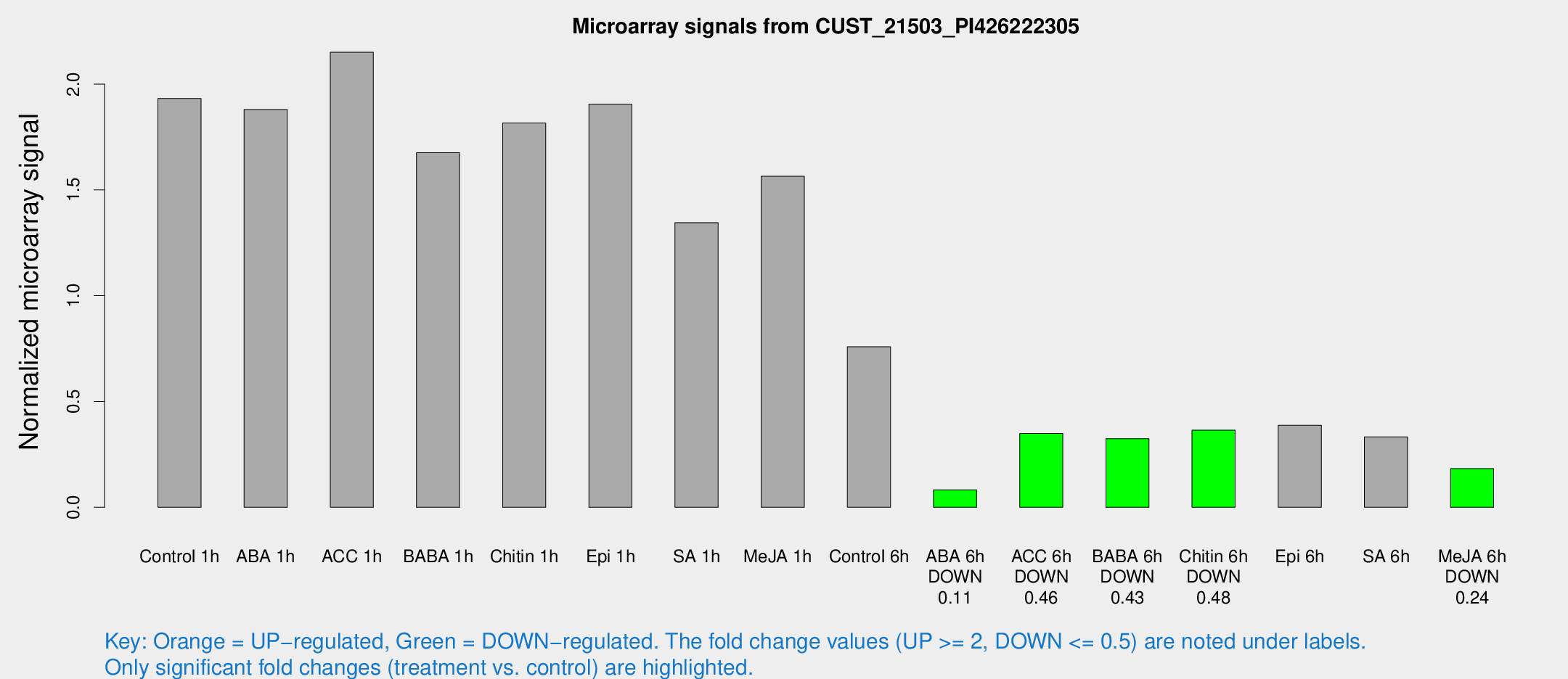
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1998.75 | 166.217 | 1.93213 | 0.11946 |
| ABA 1h | 1732.45 | 199.627 | 1.8793 | 0.36382 |
| ACC 1h | 2268.25 | 131.012 | 2.15042 | 0.171829 |
| BABA 1h | 1661.16 | 96.0888 | 1.67583 | 0.122522 |
| Chitin 1h | 1789.34 | 479.21 | 1.81588 | 0.491963 |
| Epi 1h | 1699.59 | 99.7349 | 1.90527 | 0.110058 |
| SA 1h | 1461.43 | 259.444 | 1.34497 | 0.227064 |
| Me-JA 1h | 1317.88 | 89.0721 | 1.5646 | 0.123105 |
| Control 6h | 807.127 | 144.172 | 0.758356 | 0.0736147 |
| ABA 6h | 92.7761 | 14.3003 | 0.0827135 | 0.0100905 |
| ACC 6h | 433.35 | 95.2345 | 0.348979 | 0.0464309 |
| BABA 6h | 382.776 | 62.4121 | 0.324405 | 0.0464216 |
| Chitin 6h | 403.853 | 45.5356 | 0.364288 | 0.0449932 |
| Epi 6h | 505.546 | 151.608 | 0.387523 | 0.189195 |
| SA 6h | 497.187 | 210.314 | 0.332573 | 0.471184 |
| Me-JA 6h | 214.09 | 81.3015 | 0.182852 | 0.0547437 |
Source Transcript PGSC0003DMT400070176 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G13630.1 | +1 | 0.0 | 2360 | 1190/1383 (86%) | magnesium-chelatase subunit chlH, chloroplast, putative / Mg-protoporphyrin IX chelatase, putative (CHLH) | chr5:4387567-4392082 REVERSE LENGTH=1381 |
