Probe CUST_21007_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21007_PI426222305 | JHI_St_60k_v1 | DMT400040876 | ATGTATGATCACGAAGGTCTATGACCTAGGTTGAGTCGATGTGTGATCGATCACAAAGGC |
All Microarray Probes Designed to Gene DMG400015808
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_21007_PI426222305 | JHI_St_60k_v1 | DMT400040876 | ATGTATGATCACGAAGGTCTATGACCTAGGTTGAGTCGATGTGTGATCGATCACAAAGGC |
Microarray Signals from CUST_21007_PI426222305
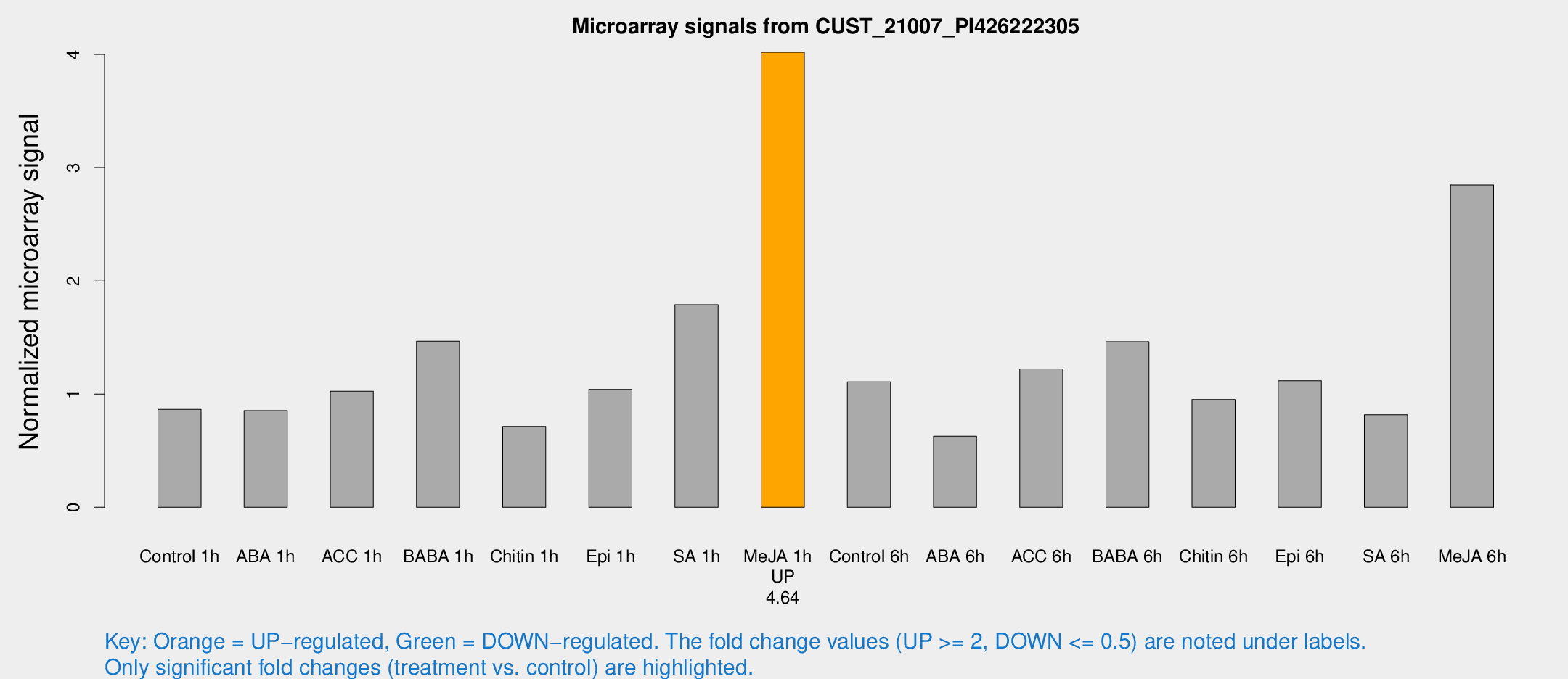
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 83.0502 | 12.0149 | 0.86545 | 0.102592 |
| ABA 1h | 72.6263 | 11.04 | 0.85466 | 0.0685049 |
| ACC 1h | 108.584 | 34.2724 | 1.02624 | 0.278324 |
| BABA 1h | 133.516 | 8.49153 | 1.46831 | 0.16655 |
| Chitin 1h | 62.6514 | 11.4263 | 0.714836 | 0.0845435 |
| Epi 1h | 85.603 | 8.57729 | 1.04176 | 0.105393 |
| SA 1h | 194.566 | 67.4211 | 1.78905 | 0.736467 |
| Me-JA 1h | 309.176 | 18.2126 | 4.01947 | 0.334951 |
| Control 6h | 120.209 | 45.2111 | 1.10972 | 0.344339 |
| ABA 6h | 65.3586 | 12.3727 | 0.627969 | 0.0996397 |
| ACC 6h | 134.871 | 20.0914 | 1.22294 | 0.276355 |
| BABA 6h | 154.689 | 10.7715 | 1.46325 | 0.0918315 |
| Chitin 6h | 95.8401 | 7.32561 | 0.952442 | 0.100157 |
| Epi 6h | 122.019 | 19.5211 | 1.11798 | 0.278946 |
| SA 6h | 76.1654 | 5.71379 | 0.817919 | 0.121348 |
| Me-JA 6h | 266.846 | 15.7984 | 2.84653 | 0.168313 |
Source Transcript PGSC0003DMT400040876 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G33150.1 | +3 | 0.0 | 660 | 364/462 (79%) | peroxisomal 3-ketoacyl-CoA thiolase 3 | chr2:14047814-14050983 REVERSE LENGTH=462 |
