Probe CUST_20916_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20916_PI426222305 | JHI_St_60k_v1 | DMT400011709 | AAATCAGAGGAAGCCTACTCAAAAGCAGGGGAAGCAAAAGAAAAAGCCTACTCAGAAGCC |
All Microarray Probes Designed to Gene DMG400004598
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20757_PI426222305 | JHI_St_60k_v1 | DMT400011708 | CTGATTCAACTATGGATGCTGCTAAGAATGCTAAAGATAAGATTACTGATACTGCTTCAG |
| CUST_20916_PI426222305 | JHI_St_60k_v1 | DMT400011709 | AAATCAGAGGAAGCCTACTCAAAAGCAGGGGAAGCAAAAGAAAAAGCCTACTCAGAAGCC |
Microarray Signals from CUST_20916_PI426222305
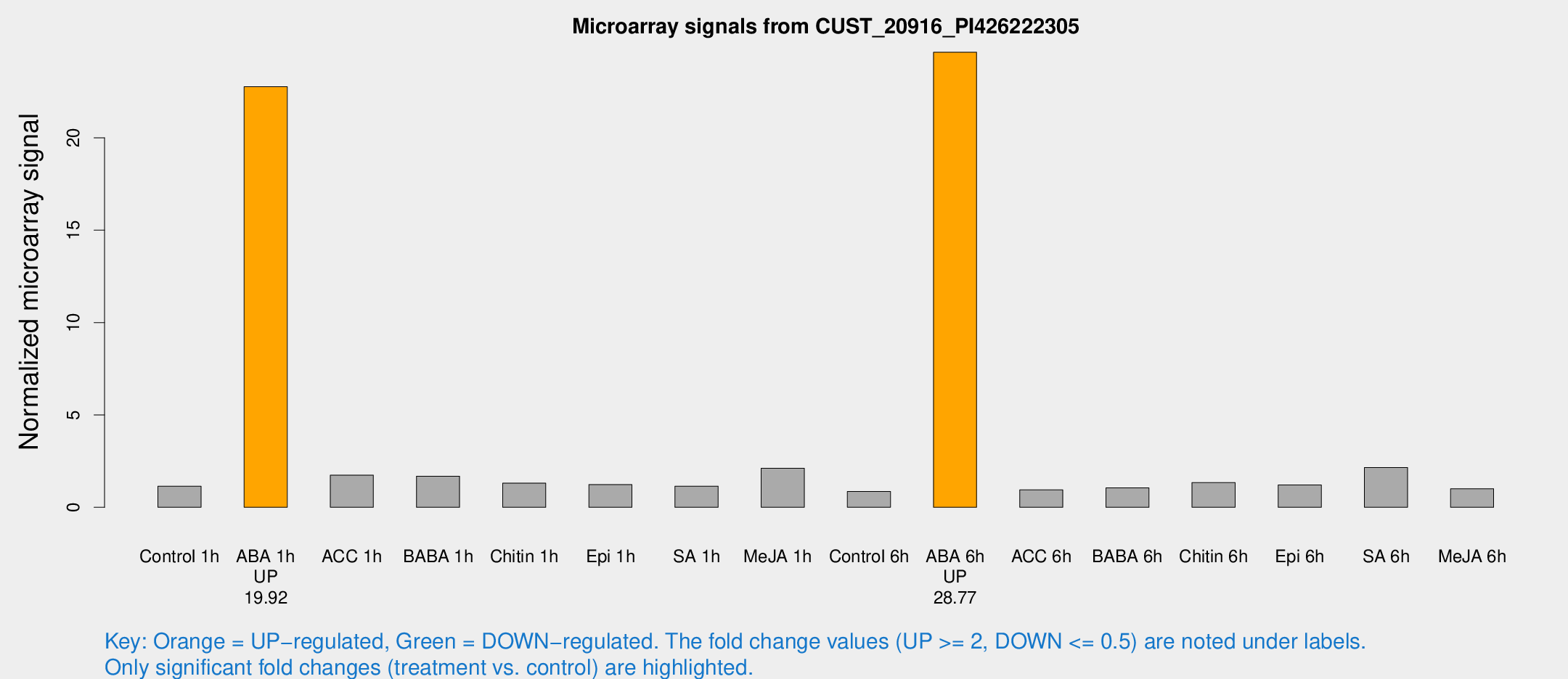
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.90005 | 3.67316 | 1.14307 | 0.508645 |
| ABA 1h | 161.113 | 17.1836 | 22.7686 | 2.56658 |
| ACC 1h | 16.0916 | 4.98642 | 1.73977 | 0.882402 |
| BABA 1h | 19.4719 | 12.5526 | 1.686 | 1.93097 |
| Chitin 1h | 10.8004 | 4.35612 | 1.30688 | 0.595563 |
| Epi 1h | 9.59511 | 3.65932 | 1.22983 | 0.584712 |
| SA 1h | 11.2043 | 5.02991 | 1.14659 | 0.527564 |
| Me-JA 1h | 19.9632 | 12.4399 | 2.11158 | 2.06907 |
| Control 6h | 6.80053 | 3.94824 | 0.856402 | 0.496517 |
| ABA 6h | 209.336 | 21.2885 | 24.6357 | 1.50345 |
| ACC 6h | 8.7028 | 4.70377 | 0.945779 | 0.502885 |
| BABA 6h | 9.64281 | 4.37525 | 1.05116 | 0.497688 |
| Chitin 6h | 13.7248 | 6.40971 | 1.33434 | 0.618649 |
| Epi 6h | 13.3667 | 6.44728 | 1.20858 | 0.566822 |
| SA 6h | 20.6008 | 8.58509 | 2.15217 | 1.59274 |
| Me-JA 6h | 8.12547 | 3.93628 | 1.0044 | 0.511654 |
Source Transcript PGSC0003DMT400011709 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
