Probe CUST_20757_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20757_PI426222305 | JHI_St_60k_v1 | DMT400011708 | CTGATTCAACTATGGATGCTGCTAAGAATGCTAAAGATAAGATTACTGATACTGCTTCAG |
All Microarray Probes Designed to Gene DMG400004598
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20916_PI426222305 | JHI_St_60k_v1 | DMT400011709 | AAATCAGAGGAAGCCTACTCAAAAGCAGGGGAAGCAAAAGAAAAAGCCTACTCAGAAGCC |
| CUST_20757_PI426222305 | JHI_St_60k_v1 | DMT400011708 | CTGATTCAACTATGGATGCTGCTAAGAATGCTAAAGATAAGATTACTGATACTGCTTCAG |
Microarray Signals from CUST_20757_PI426222305
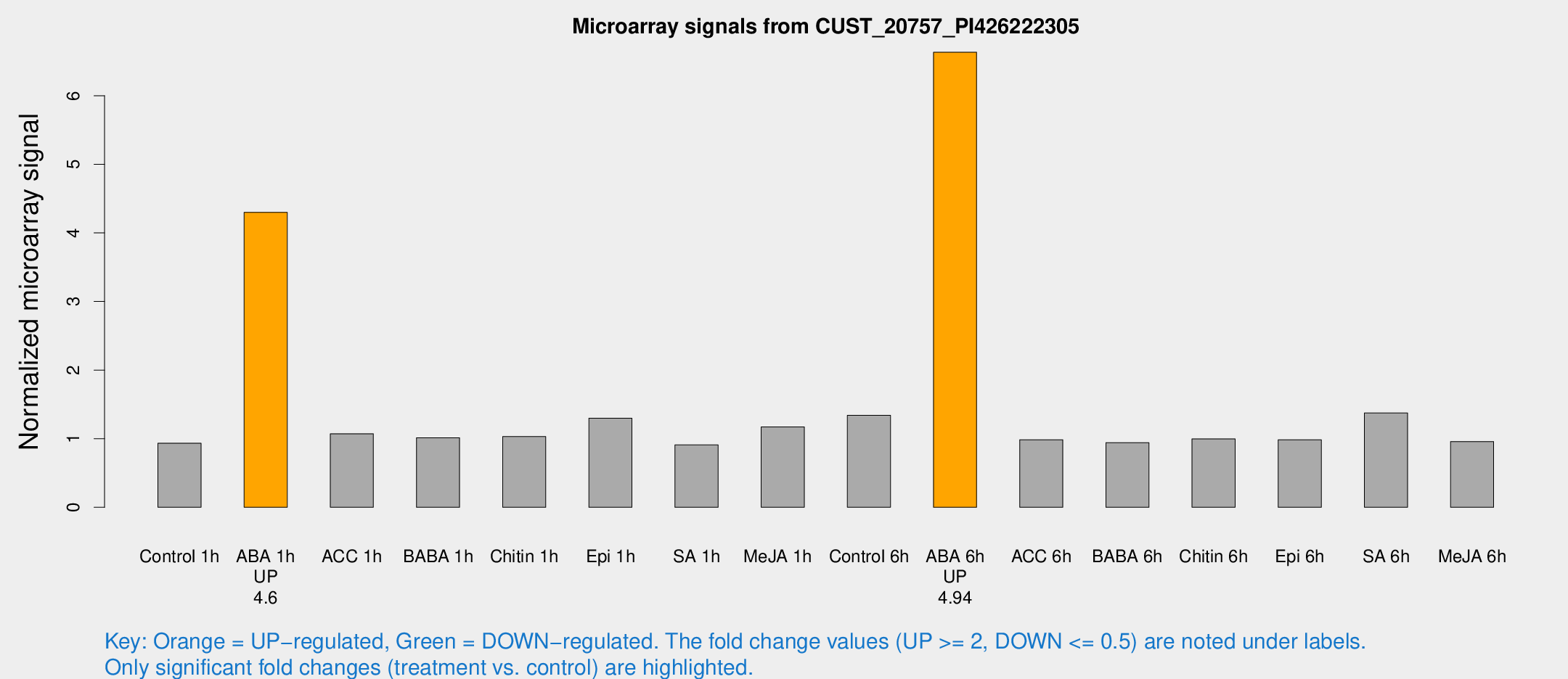
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.68597 | 3.29951 | 0.934843 | 0.541809 |
| ABA 1h | 23.5405 | 3.46313 | 4.29957 | 0.664884 |
| ACC 1h | 6.80378 | 4.01326 | 1.07157 | 0.62052 |
| BABA 1h | 5.94611 | 3.44904 | 1.01458 | 0.588005 |
| Chitin 1h | 5.65058 | 3.28061 | 1.03173 | 0.597461 |
| Epi 1h | 7.26175 | 3.24783 | 1.29865 | 0.643484 |
| SA 1h | 5.68039 | 3.29153 | 0.909843 | 0.526962 |
| Me-JA 1h | 5.82997 | 3.38552 | 1.17253 | 0.679119 |
| Control 6h | 8.60962 | 3.39308 | 1.34159 | 0.591019 |
| ABA 6h | 44.7907 | 8.73948 | 6.63333 | 1.24456 |
| ACC 6h | 7.04394 | 4.19167 | 0.984793 | 0.570264 |
| BABA 6h | 6.42192 | 3.72661 | 0.942376 | 0.545712 |
| Chitin 6h | 6.45143 | 3.74084 | 0.997207 | 0.578014 |
| Epi 6h | 6.79236 | 3.96185 | 0.984929 | 0.570608 |
| SA 6h | 9.27127 | 3.57739 | 1.37505 | 0.72132 |
| Me-JA 6h | 5.80743 | 3.36671 | 0.95887 | 0.555243 |
Source Transcript PGSC0003DMT400011708 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G36600.1 | +3 | 2e-06 | 49 | 26/77 (34%) | Late embryogenesis abundant (LEA) protein | chr4:17263666-17264968 FORWARD LENGTH=353 |
