Probe CUST_20436_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20436_PI426222305 | JHI_St_60k_v1 | DMT400068335 | TTTGTGTCAACTTTGGCTGTTTGCACACTGCAAACATTATCTCTGATGATGAATGGAATT |
All Microarray Probes Designed to Gene DMG400026571
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20436_PI426222305 | JHI_St_60k_v1 | DMT400068335 | TTTGTGTCAACTTTGGCTGTTTGCACACTGCAAACATTATCTCTGATGATGAATGGAATT |
Microarray Signals from CUST_20436_PI426222305
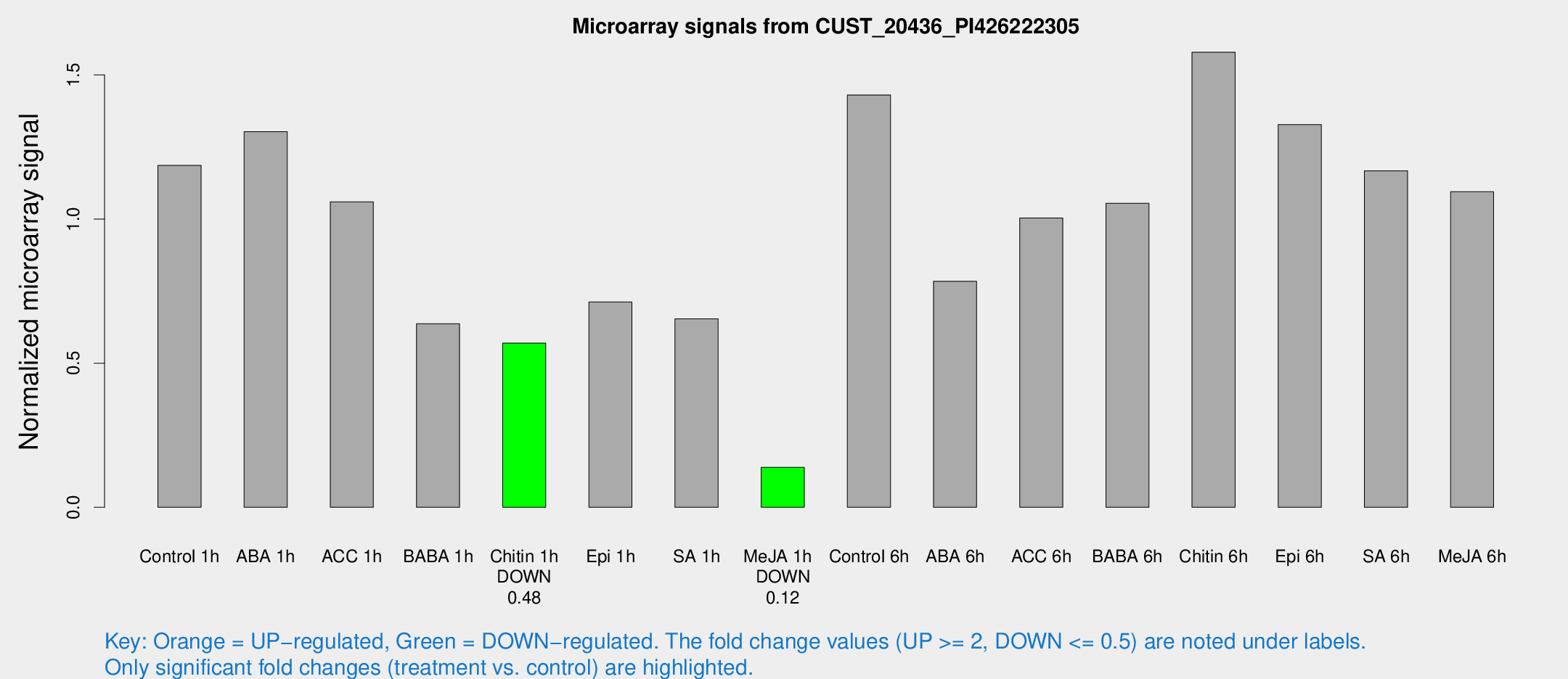
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1341.23 | 150.371 | 1.18606 | 0.0704028 |
| ABA 1h | 1288.34 | 74.565 | 1.30318 | 0.119468 |
| ACC 1h | 1357.39 | 393.128 | 1.05973 | 0.31113 |
| BABA 1h | 691.931 | 67.377 | 0.636907 | 0.0368861 |
| Chitin 1h | 573.716 | 35.715 | 0.569632 | 0.0330113 |
| Epi 1h | 695.855 | 77.3178 | 0.712281 | 0.083906 |
| SA 1h | 769.083 | 125.184 | 0.653894 | 0.0964809 |
| Me-JA 1h | 126.601 | 7.90478 | 0.138742 | 0.0141932 |
| Control 6h | 1650.84 | 273.691 | 1.43064 | 0.124175 |
| ABA 6h | 937.395 | 78.1757 | 0.784495 | 0.0734242 |
| ACC 6h | 1298.34 | 133.705 | 1.00357 | 0.091389 |
| BABA 6h | 1349.65 | 216.672 | 1.05496 | 0.167895 |
| Chitin 6h | 1878.92 | 108.745 | 1.57896 | 0.106036 |
| Epi 6h | 1683.29 | 145.091 | 1.3276 | 0.208311 |
| SA 6h | 1319.47 | 191.36 | 1.16761 | 0.0674819 |
| Me-JA 6h | 1359.16 | 450.563 | 1.09478 | 0.28727 |
Source Transcript PGSC0003DMT400068335 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
