Probe CUST_20178_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20178_PI426222305 | JHI_St_60k_v1 | DMT400068922 | TACTTTATTTCAAGGGACGTATCACGCGCCGCAATGGTTACTATTATCGAGGACCGTATC |
All Microarray Probes Designed to Gene DMG400026807
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20178_PI426222305 | JHI_St_60k_v1 | DMT400068922 | TACTTTATTTCAAGGGACGTATCACGCGCCGCAATGGTTACTATTATCGAGGACCGTATC |
Microarray Signals from CUST_20178_PI426222305
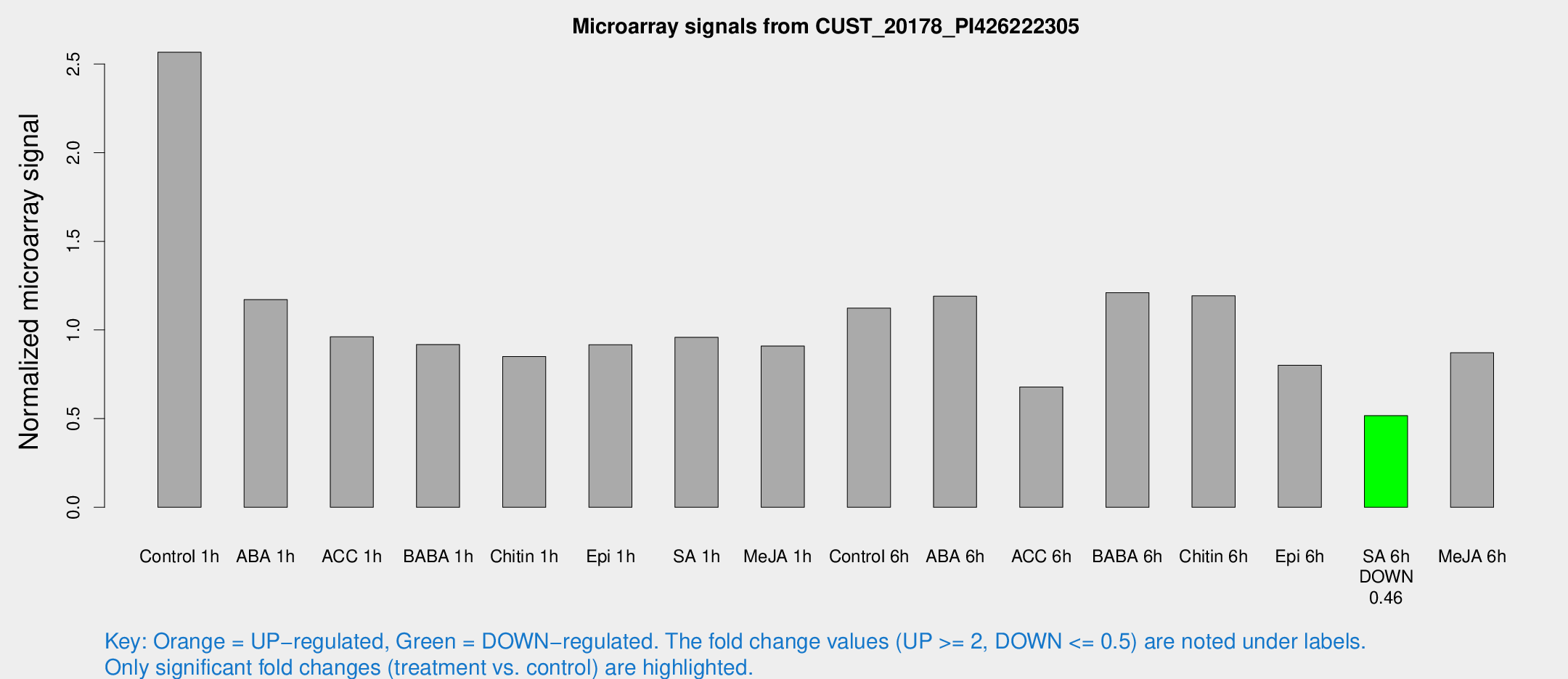
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 293.969 | 253.527 | 2.56617 | 5.36152 |
| ABA 1h | 44.1448 | 4.37887 | 1.17098 | 0.116132 |
| ACC 1h | 43.5081 | 8.27141 | 0.961555 | 0.175274 |
| BABA 1h | 37.9692 | 4.52743 | 0.918153 | 0.111092 |
| Chitin 1h | 33.0065 | 4.19 | 0.850533 | 0.107846 |
| Epi 1h | 33.9663 | 4.11529 | 0.917138 | 0.112638 |
| SA 1h | 42.3089 | 4.26651 | 0.958493 | 0.121233 |
| Me-JA 1h | 31.7709 | 4.20573 | 0.909487 | 0.121432 |
| Control 6h | 51.0742 | 11.6353 | 1.12306 | 0.200314 |
| ABA 6h | 55.0024 | 8.00743 | 1.19033 | 0.118483 |
| ACC 6h | 34.9788 | 7.34985 | 0.678836 | 0.148433 |
| BABA 6h | 59.0925 | 8.47254 | 1.21042 | 0.146756 |
| Chitin 6h | 54.1612 | 5.43703 | 1.19236 | 0.119836 |
| Epi 6h | 41.1713 | 9.49146 | 0.800987 | 0.298917 |
| SA 6h | 21.9206 | 4.35014 | 0.516974 | 0.103776 |
| Me-JA 6h | 43.8841 | 16.2673 | 0.871831 | 0.336942 |
Source Transcript PGSC0003DMT400068922 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
