Probe CUST_20005_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20005_PI426222305 | JHI_St_60k_v1 | DMT400049193 | AGAACAAAGGGTCCTTGTGGAAAATCTCAAGTTGAATCACTAAAATCGCCAAGATAAATA |
All Microarray Probes Designed to Gene DMG400019120
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_20005_PI426222305 | JHI_St_60k_v1 | DMT400049193 | AGAACAAAGGGTCCTTGTGGAAAATCTCAAGTTGAATCACTAAAATCGCCAAGATAAATA |
Microarray Signals from CUST_20005_PI426222305
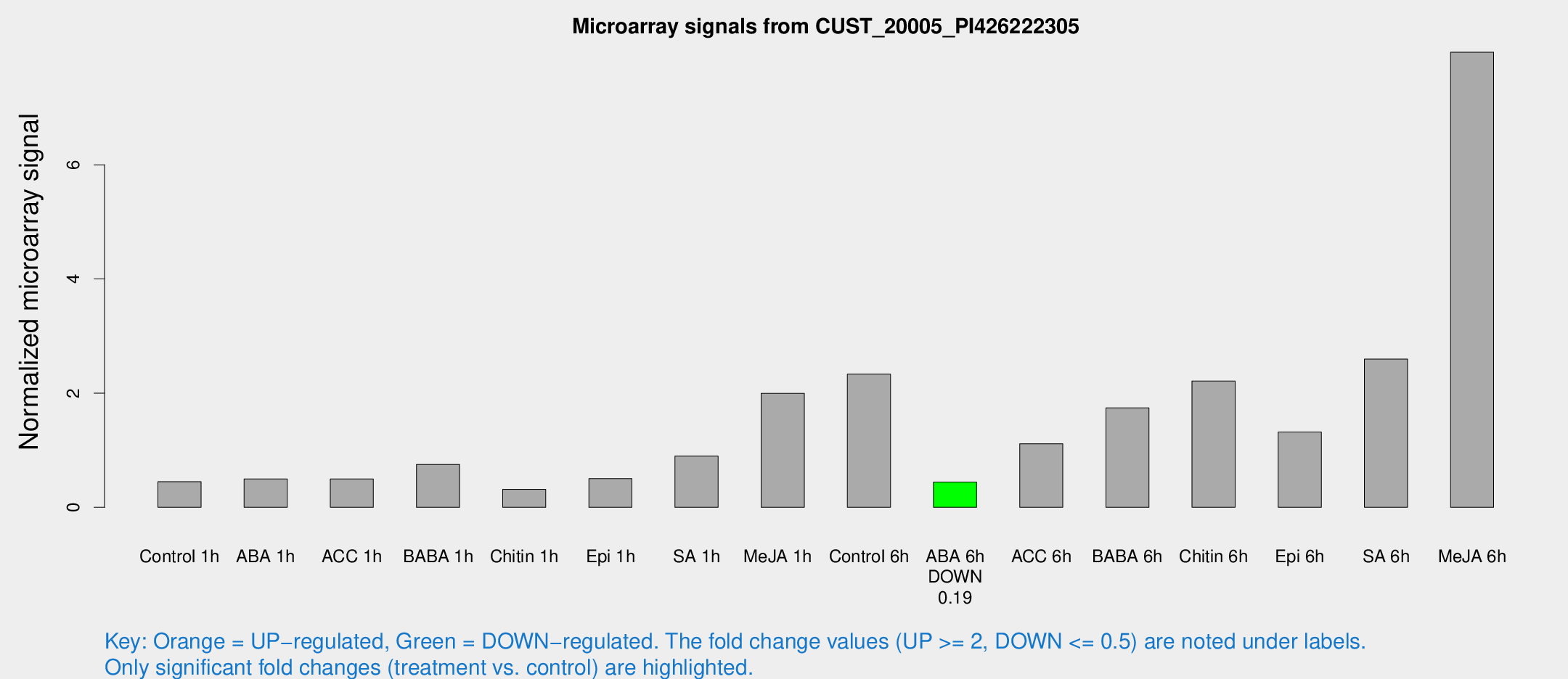
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 45.1864 | 15.1061 | 0.447965 | 0.116572 |
| ABA 1h | 41.7312 | 8.25526 | 0.495053 | 0.0907115 |
| ACC 1h | 60.3434 | 30.4813 | 0.495182 | 0.270563 |
| BABA 1h | 73.9478 | 25.3809 | 0.751238 | 0.207892 |
| Chitin 1h | 29.7904 | 11.923 | 0.313999 | 0.106262 |
| Epi 1h | 42.401 | 10.3819 | 0.50164 | 0.133155 |
| SA 1h | 85.3518 | 11.1826 | 0.895943 | 0.0675684 |
| Me-JA 1h | 168.686 | 63.3456 | 1.99516 | 0.599282 |
| Control 6h | 244.225 | 93.5492 | 2.33219 | 0.847233 |
| ABA 6h | 46.6391 | 12.7497 | 0.442634 | 0.154529 |
| ACC 6h | 118.468 | 15.993 | 1.11345 | 0.240822 |
| BABA 6h | 177.848 | 10.9077 | 1.74244 | 0.106705 |
| Chitin 6h | 221.937 | 42.9354 | 2.21078 | 0.409269 |
| Epi 6h | 136.555 | 11.9012 | 1.31981 | 0.220669 |
| SA 6h | 236.216 | 23.2576 | 2.5949 | 0.154764 |
| Me-JA 6h | 726.165 | 49.1999 | 7.97127 | 0.963503 |
Source Transcript PGSC0003DMT400049193 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G55970.1 | +1 | 8e-84 | 256 | 124/186 (67%) | jasmonate-regulated gene 21 | chr3:20766970-20769264 REVERSE LENGTH=363 |
