Probe CUST_19941_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19941_PI426222305 | JHI_St_60k_v1 | DMT400049192 | GGAAGTTACTGTGTATATATGCCAATTGGTAAAACATAACACAGTAAATACCCTATGTCC |
All Microarray Probes Designed to Gene DMG400019119
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19941_PI426222305 | JHI_St_60k_v1 | DMT400049192 | GGAAGTTACTGTGTATATATGCCAATTGGTAAAACATAACACAGTAAATACCCTATGTCC |
Microarray Signals from CUST_19941_PI426222305
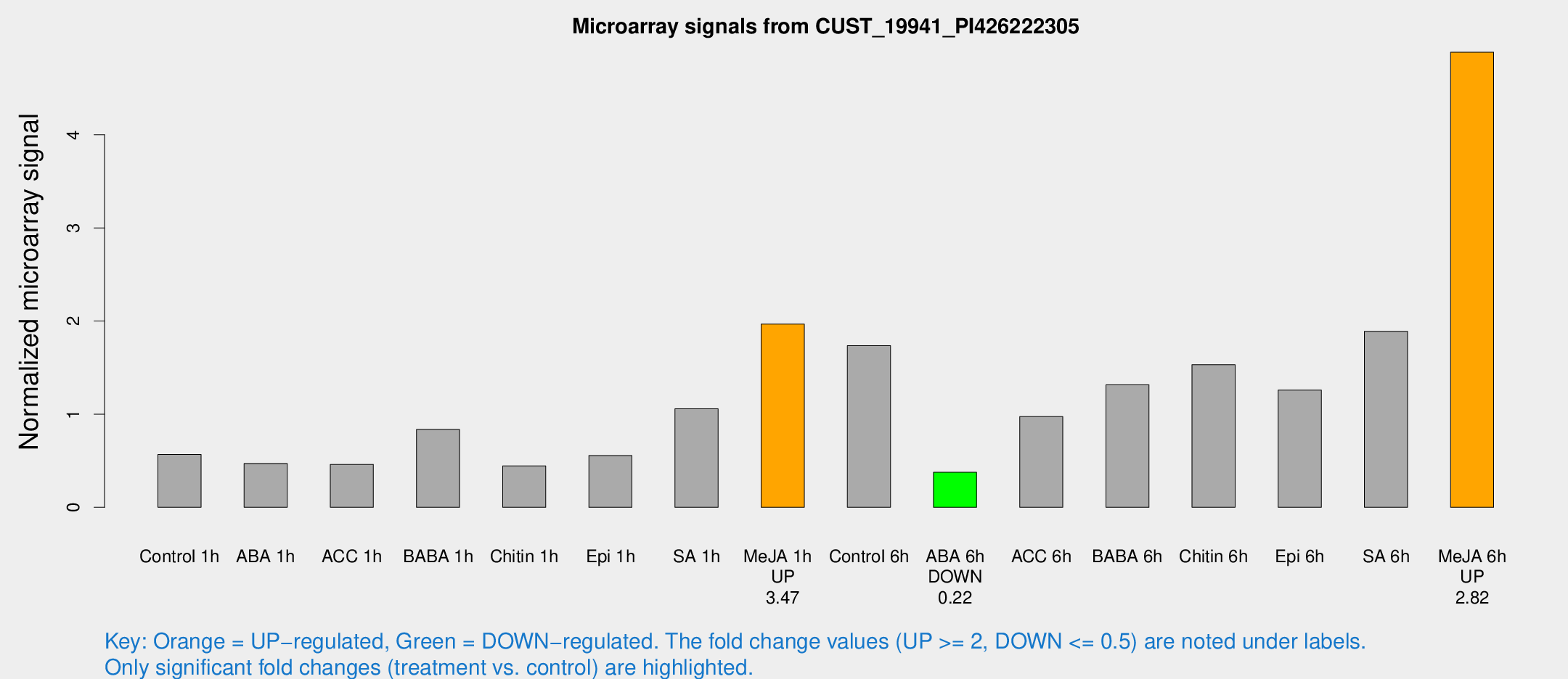
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 170.167 | 23.8525 | 0.566727 | 0.0456892 |
| ABA 1h | 154.397 | 68.9589 | 0.470521 | 0.232098 |
| ACC 1h | 182.66 | 72.8056 | 0.45949 | 0.291927 |
| BABA 1h | 255.693 | 68.2448 | 0.835181 | 0.164194 |
| Chitin 1h | 118.15 | 7.55058 | 0.443876 | 0.0279213 |
| Epi 1h | 145.378 | 22.312 | 0.555417 | 0.0876121 |
| SA 1h | 343.917 | 94.1799 | 1.0586 | 0.315659 |
| Me-JA 1h | 490.658 | 96.4969 | 1.96741 | 0.242783 |
| Control 6h | 577.578 | 202.597 | 1.73501 | 0.481773 |
| ABA 6h | 119.064 | 11.8263 | 0.375994 | 0.0497155 |
| ACC 6h | 346.48 | 70.2015 | 0.97443 | 0.234508 |
| BABA 6h | 437.442 | 37.4007 | 1.31592 | 0.0903387 |
| Chitin 6h | 489.668 | 68.2063 | 1.53208 | 0.206195 |
| Epi 6h | 432.469 | 73.8185 | 1.25821 | 0.32759 |
| SA 6h | 554.901 | 46.5122 | 1.88932 | 0.292317 |
| Me-JA 6h | 1460.68 | 202.411 | 4.88697 | 0.744156 |
Source Transcript PGSC0003DMT400049192 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G55970.1 | +1 | 1e-155 | 451 | 222/365 (61%) | jasmonate-regulated gene 21 | chr3:20766970-20769264 REVERSE LENGTH=363 |
