Probe CUST_19932_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19932_PI426222305 | JHI_St_60k_v1 | DMT400061156 | GGGGATTTTGATTCTCCCTATTATAATTTCCTTCCCCGCCCGCATCTATGTAATAAAAAA |
All Microarray Probes Designed to Gene DMG400023798
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19932_PI426222305 | JHI_St_60k_v1 | DMT400061156 | GGGGATTTTGATTCTCCCTATTATAATTTCCTTCCCCGCCCGCATCTATGTAATAAAAAA |
Microarray Signals from CUST_19932_PI426222305
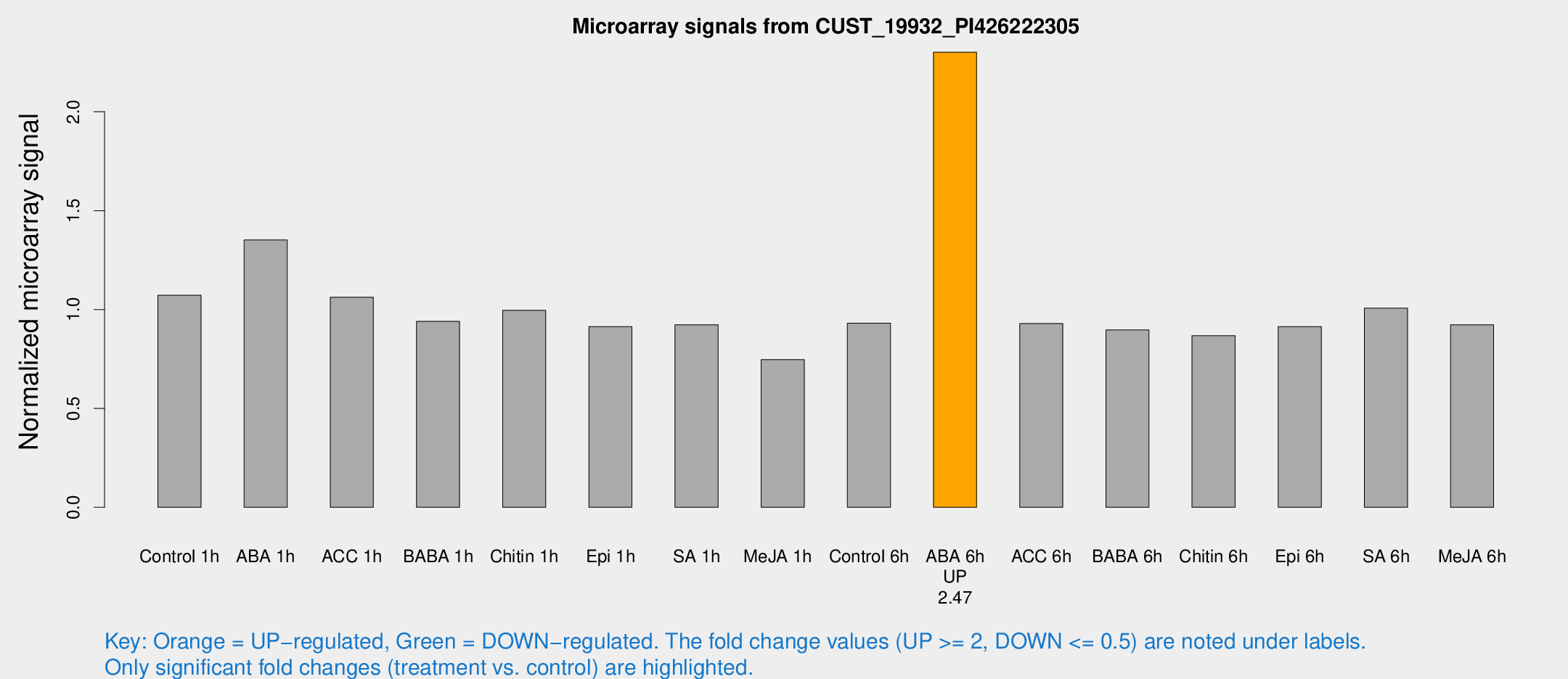
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1851.96 | 107.26 | 1.0722 | 0.0619337 |
| ABA 1h | 2061.99 | 119.189 | 1.35195 | 0.119548 |
| ACC 1h | 1907.47 | 225.856 | 1.06257 | 0.0613985 |
| BABA 1h | 1583.48 | 191.885 | 0.940127 | 0.0732916 |
| Chitin 1h | 1551.38 | 114.909 | 0.99596 | 0.0770262 |
| Epi 1h | 1369.42 | 96.6039 | 0.913731 | 0.0626312 |
| SA 1h | 1643.6 | 127.506 | 0.923065 | 0.115764 |
| Me-JA 1h | 1051.76 | 60.9066 | 0.746688 | 0.0940673 |
| Control 6h | 1643.36 | 232.64 | 0.930968 | 0.0661732 |
| ABA 6h | 4228.27 | 244.897 | 2.30115 | 0.132871 |
| ACC 6h | 1860.07 | 200.897 | 0.929256 | 0.137575 |
| BABA 6h | 1729.48 | 99.9333 | 0.89673 | 0.0518105 |
| Chitin 6h | 1609.3 | 175.051 | 0.867725 | 0.0922058 |
| Epi 6h | 1775.69 | 102.602 | 0.914085 | 0.0707193 |
| SA 6h | 1746.77 | 241.715 | 1.00677 | 0.10799 |
| Me-JA 6h | 1594.4 | 133.264 | 0.923095 | 0.0691943 |
Source Transcript PGSC0003DMT400061156 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G41475.1 | +1 | 5e-53 | 174 | 90/162 (56%) | Embryo-specific protein 3, (ATS3) | chr2:17295259-17296329 REVERSE LENGTH=179 |
