Probe CUST_19849_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19849_PI426222305 | JHI_St_60k_v1 | DMT400064092 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
All Microarray Probes Designed to Gene DMG400024906
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19742_PI426222305 | JHI_St_60k_v1 | DMT400064097 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19670_PI426222305 | JHI_St_60k_v1 | DMT400064090 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19849_PI426222305 | JHI_St_60k_v1 | DMT400064092 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19766_PI426222305 | JHI_St_60k_v1 | DMT400064091 | GGGAAGAACTTGTCTGATTCTGATACATTGGTATGATCTGCTAATCTTACATGCTTAAAT |
| CUST_19684_PI426222305 | JHI_St_60k_v1 | DMT400064093 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19937_PI426222305 | JHI_St_60k_v1 | DMT400064096 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19855_PI426222305 | JHI_St_60k_v1 | DMT400064095 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
Microarray Signals from CUST_19849_PI426222305
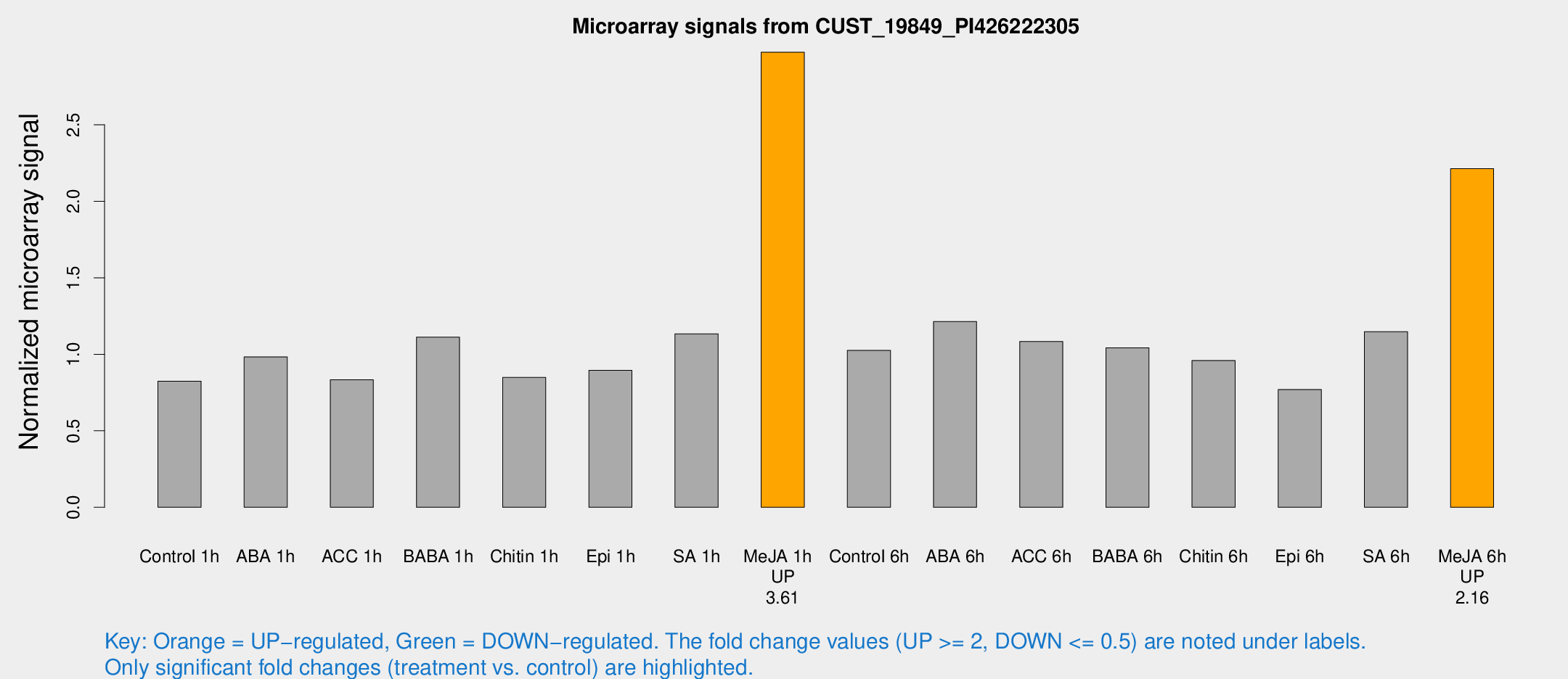
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 192.106 | 25.1847 | 0.823599 | 0.0837104 |
| ABA 1h | 210.119 | 43.5277 | 0.982658 | 0.191052 |
| ACC 1h | 215.523 | 68.8617 | 0.833773 | 0.231422 |
| BABA 1h | 253.829 | 43.1302 | 1.11173 | 0.0945293 |
| Chitin 1h | 177.476 | 19.909 | 0.848932 | 0.0695964 |
| Epi 1h | 178.436 | 10.9552 | 0.895635 | 0.0549656 |
| SA 1h | 287.512 | 80.9048 | 1.13412 | 0.305345 |
| Me-JA 1h | 567.332 | 75.5268 | 2.97431 | 0.172936 |
| Control 6h | 244.692 | 43.7141 | 1.0253 | 0.107575 |
| ABA 6h | 317.288 | 76.4604 | 1.21435 | 0.279206 |
| ACC 6h | 286.238 | 17.7931 | 1.08356 | 0.0927061 |
| BABA 6h | 273.936 | 39.2788 | 1.04277 | 0.121038 |
| Chitin 6h | 237.425 | 26.1403 | 0.959693 | 0.121013 |
| Epi 6h | 203.481 | 30.7507 | 0.769886 | 0.106906 |
| SA 6h | 265.068 | 35.1521 | 1.14789 | 0.296189 |
| Me-JA 6h | 512.262 | 56.1153 | 2.2135 | 0.128992 |
Source Transcript PGSC0003DMT400064092 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18530.1 | +3 | 3e-63 | 212 | 100/135 (74%) | Protein of unknown function (DUF707) | chr4:10217749-10220285 FORWARD LENGTH=389 |
