Probe CUST_19670_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19670_PI426222305 | JHI_St_60k_v1 | DMT400064090 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
All Microarray Probes Designed to Gene DMG400024906
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19684_PI426222305 | JHI_St_60k_v1 | DMT400064093 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19849_PI426222305 | JHI_St_60k_v1 | DMT400064092 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19855_PI426222305 | JHI_St_60k_v1 | DMT400064095 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19670_PI426222305 | JHI_St_60k_v1 | DMT400064090 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19742_PI426222305 | JHI_St_60k_v1 | DMT400064097 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19937_PI426222305 | JHI_St_60k_v1 | DMT400064096 | GCAAGATCAATGTTGGGTTGATCCATTTCAAAGTCAAGAAAAAAGAAAAACAGGCACATA |
| CUST_19766_PI426222305 | JHI_St_60k_v1 | DMT400064091 | GGGAAGAACTTGTCTGATTCTGATACATTGGTATGATCTGCTAATCTTACATGCTTAAAT |
Microarray Signals from CUST_19670_PI426222305
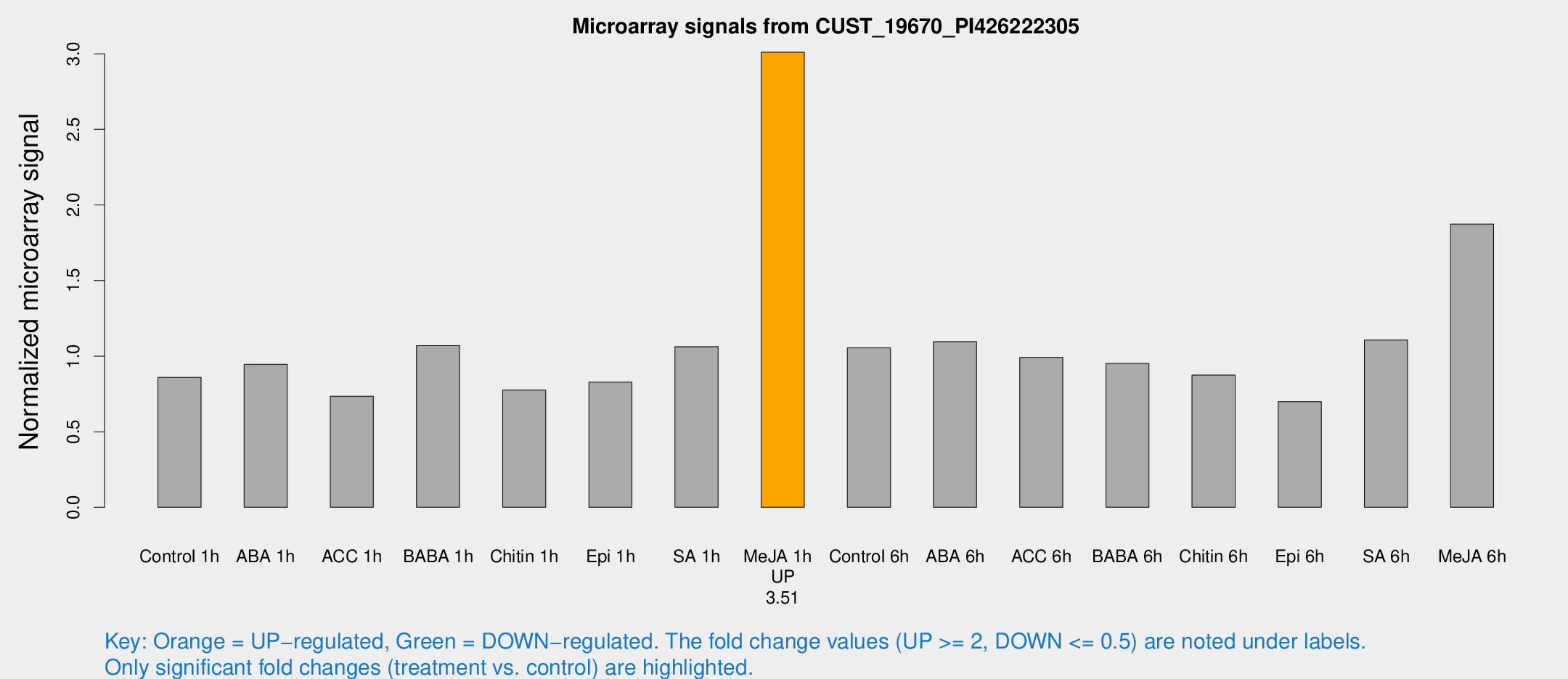
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 220.87 | 19.5901 | 0.85884 | 0.0753251 |
| ABA 1h | 223.032 | 42.6901 | 0.945713 | 0.160742 |
| ACC 1h | 216.816 | 78.8775 | 0.734526 | 0.246944 |
| BABA 1h | 266.254 | 28.4091 | 1.0698 | 0.0636631 |
| Chitin 1h | 179.402 | 16.8667 | 0.775483 | 0.0864614 |
| Epi 1h | 183.61 | 11.9914 | 0.828003 | 0.0549022 |
| SA 1h | 306.656 | 99.3329 | 1.06238 | 0.334411 |
| Me-JA 1h | 638.21 | 82.0024 | 3.01034 | 0.174723 |
| Control 6h | 279.973 | 50.1844 | 1.05456 | 0.117949 |
| ABA 6h | 317.945 | 75.112 | 1.09625 | 0.248858 |
| ACC 6h | 291.763 | 20.6652 | 0.991738 | 0.0918848 |
| BABA 6h | 278.31 | 40.5624 | 0.951328 | 0.116913 |
| Chitin 6h | 243.127 | 37.2757 | 0.874429 | 0.153358 |
| Epi 6h | 203.124 | 20.8502 | 0.699118 | 0.0897502 |
| SA 6h | 291.941 | 58.6649 | 1.10625 | 0.382275 |
| Me-JA 6h | 482.999 | 57.1242 | 1.87276 | 0.109103 |
Source Transcript PGSC0003DMT400064090 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G18530.1 | +1 | 8e-122 | 362 | 199/416 (48%) | Protein of unknown function (DUF707) | chr4:10217749-10220285 FORWARD LENGTH=389 |
