Probe CUST_19604_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19604_PI426222305 | JHI_St_60k_v1 | DMT400023321 | GAAGCCTTAGTTCAGGTACGAATTTCGGTGTTTTGACCATTTTTTATTGTGAAATTTGAG |
All Microarray Probes Designed to Gene DMG400009033
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19604_PI426222305 | JHI_St_60k_v1 | DMT400023321 | GAAGCCTTAGTTCAGGTACGAATTTCGGTGTTTTGACCATTTTTTATTGTGAAATTTGAG |
| CUST_19574_PI426222305 | JHI_St_60k_v1 | DMT400023322 | GGTGAGACTAATTTTCATTGCCTTGTGCCTCTCTTCATTTTGTAATAATGGGAAGAACAC |
Microarray Signals from CUST_19604_PI426222305
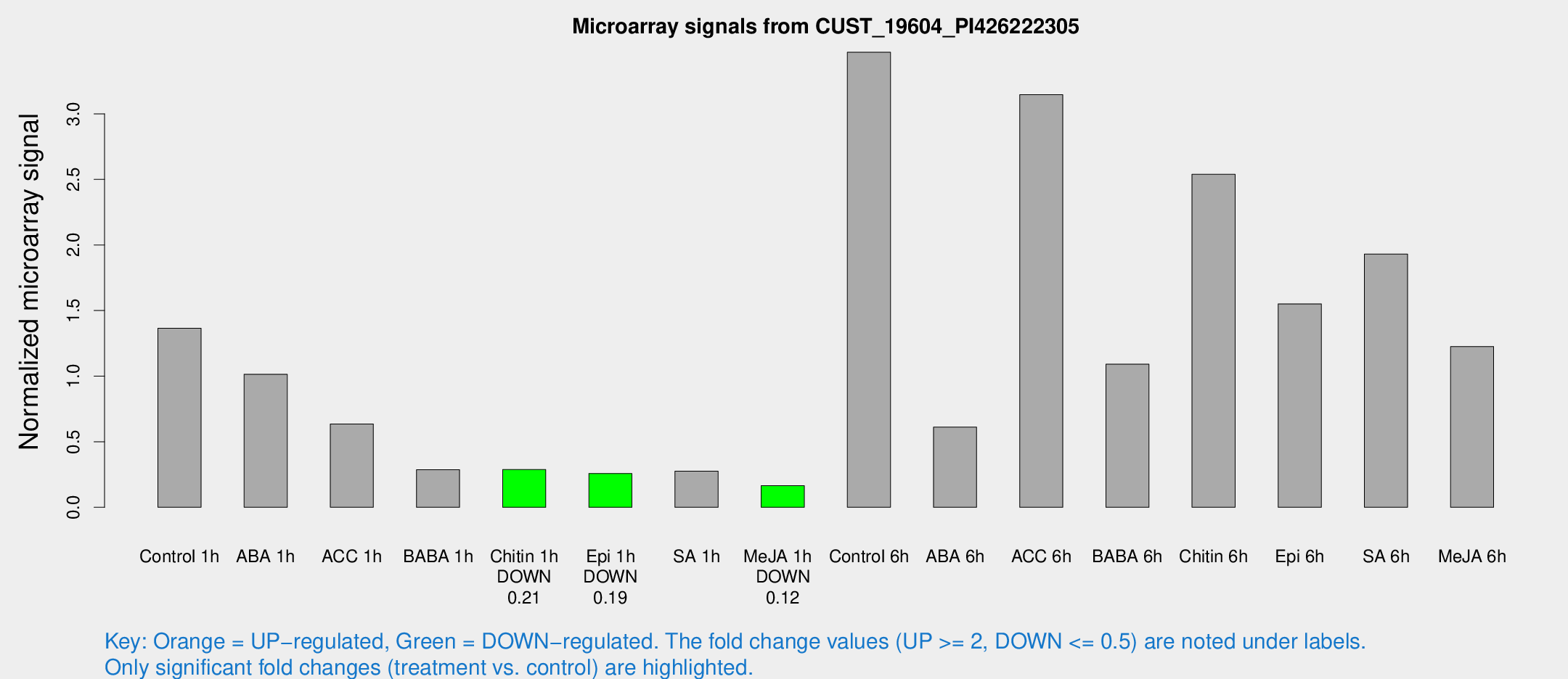
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 65.8252 | 12.3152 | 1.36553 | 0.198387 |
| ABA 1h | 57.5509 | 32.8644 | 1.01459 | 0.711701 |
| ACC 1h | 36.8375 | 12.8674 | 0.634558 | 0.304913 |
| BABA 1h | 14.1724 | 4.42782 | 0.286721 | 0.1379 |
| Chitin 1h | 13.4923 | 4.0996 | 0.287568 | 0.099921 |
| Epi 1h | 11.1688 | 3.6483 | 0.257464 | 0.099523 |
| SA 1h | 17.3784 | 9.22866 | 0.276094 | 0.170564 |
| Me-JA 1h | 6.26773 | 3.64432 | 0.165319 | 0.0958848 |
| Control 6h | 176.282 | 55.2444 | 3.46965 | 1.07008 |
| ABA 6h | 33.2992 | 9.10599 | 0.610972 | 0.271045 |
| ACC 6h | 168.015 | 11.6395 | 3.14619 | 0.456933 |
| BABA 6h | 64.2852 | 21.6573 | 1.09118 | 0.472139 |
| Chitin 6h | 127.782 | 17.2553 | 2.53879 | 0.377102 |
| Epi 6h | 81.0879 | 6.33359 | 1.55112 | 0.163332 |
| SA 6h | 90.4182 | 13.8284 | 1.93077 | 0.261674 |
| Me-JA 6h | 75.5596 | 33.5023 | 1.22574 | 0.948463 |
Source Transcript PGSC0003DMT400023321 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G47460.1 | +1 | 3e-73 | 231 | 108/133 (81%) | myb domain protein 12 | chr2:19476438-19479242 FORWARD LENGTH=371 |
