Probe CUST_19574_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19574_PI426222305 | JHI_St_60k_v1 | DMT400023322 | GGTGAGACTAATTTTCATTGCCTTGTGCCTCTCTTCATTTTGTAATAATGGGAAGAACAC |
All Microarray Probes Designed to Gene DMG400009033
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19574_PI426222305 | JHI_St_60k_v1 | DMT400023322 | GGTGAGACTAATTTTCATTGCCTTGTGCCTCTCTTCATTTTGTAATAATGGGAAGAACAC |
| CUST_19604_PI426222305 | JHI_St_60k_v1 | DMT400023321 | GAAGCCTTAGTTCAGGTACGAATTTCGGTGTTTTGACCATTTTTTATTGTGAAATTTGAG |
Microarray Signals from CUST_19574_PI426222305
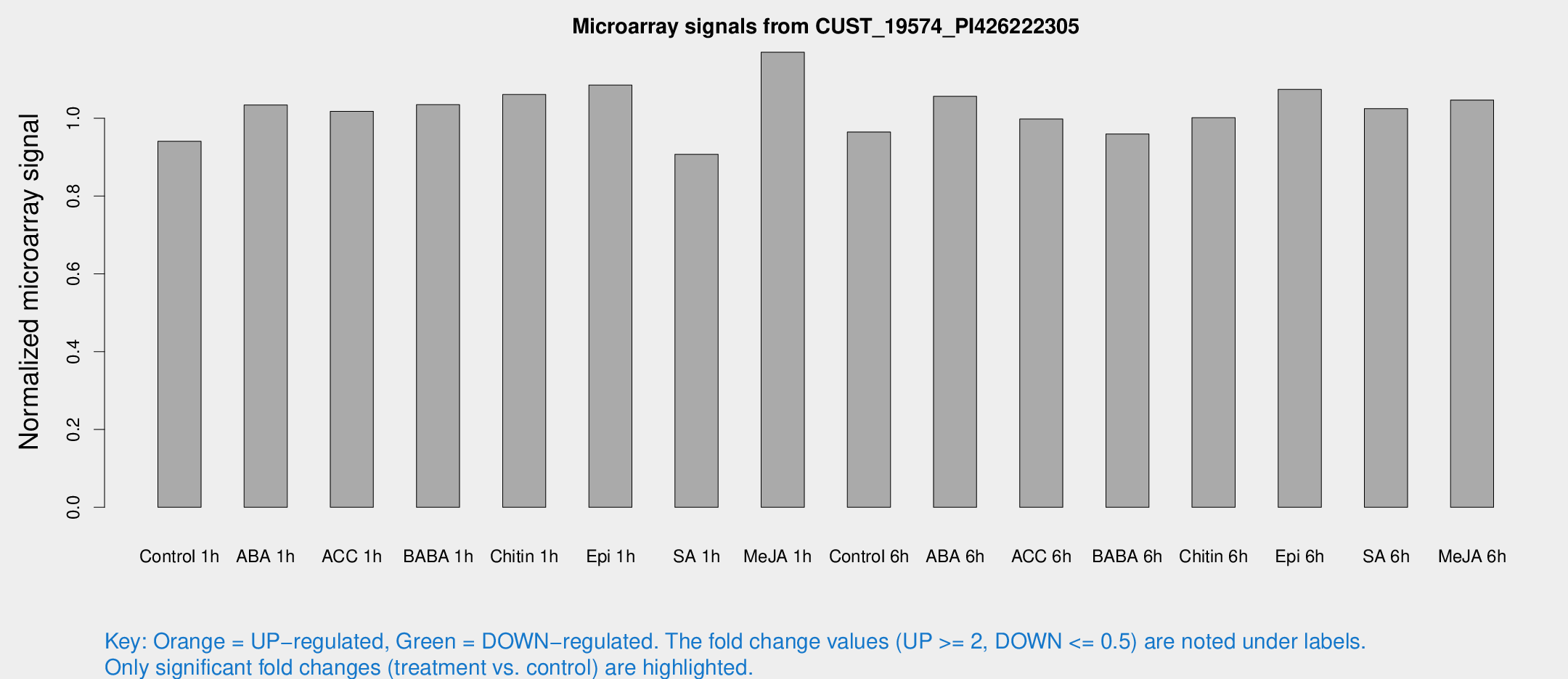
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.16381 | 2.99335 | 0.941153 | 0.53856 |
| ABA 1h | 4.99047 | 2.89272 | 1.03435 | 0.588882 |
| ACC 1h | 5.81983 | 3.38333 | 1.01783 | 0.589403 |
| BABA 1h | 5.54087 | 3.15188 | 1.03502 | 0.587795 |
| Chitin 1h | 5.16897 | 3.00612 | 1.06109 | 0.598034 |
| Epi 1h | 5.10455 | 2.96171 | 1.08549 | 0.614788 |
| SA 1h | 5.17034 | 2.9962 | 0.907344 | 0.525112 |
| Me-JA 1h | 5.29557 | 3.07786 | 1.16988 | 0.674649 |
| Control 6h | 5.36461 | 3.11237 | 0.964802 | 0.559634 |
| ABA 6h | 6.28171 | 3.22037 | 1.05637 | 0.553241 |
| ACC 6h | 6.48109 | 3.81132 | 0.998252 | 0.573526 |
| BABA 6h | 5.97199 | 3.46618 | 0.959667 | 0.555725 |
| Chitin 6h | 5.91602 | 3.43209 | 1.00149 | 0.580673 |
| Epi 6h | 6.80722 | 3.60511 | 1.07433 | 0.572624 |
| SA 6h | 5.62514 | 3.25877 | 1.02482 | 0.593453 |
| Me-JA 6h | 5.84175 | 3.11244 | 1.04696 | 0.565722 |
Source Transcript PGSC0003DMT400023322 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G47460.1 | +2 | 5e-71 | 231 | 108/133 (81%) | myb domain protein 12 | chr2:19476438-19479242 FORWARD LENGTH=371 |
