Probe CUST_19492_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19492_PI426222305 | JHI_St_60k_v1 | DMT400023192 | TTATTTTCGCTTGCTTTGTTAGTCCCAATATGGTGAGACTTCTCGCCAATGGAGTCACAG |
All Microarray Probes Designed to Gene DMG400008984
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19492_PI426222305 | JHI_St_60k_v1 | DMT400023192 | TTATTTTCGCTTGCTTTGTTAGTCCCAATATGGTGAGACTTCTCGCCAATGGAGTCACAG |
Microarray Signals from CUST_19492_PI426222305
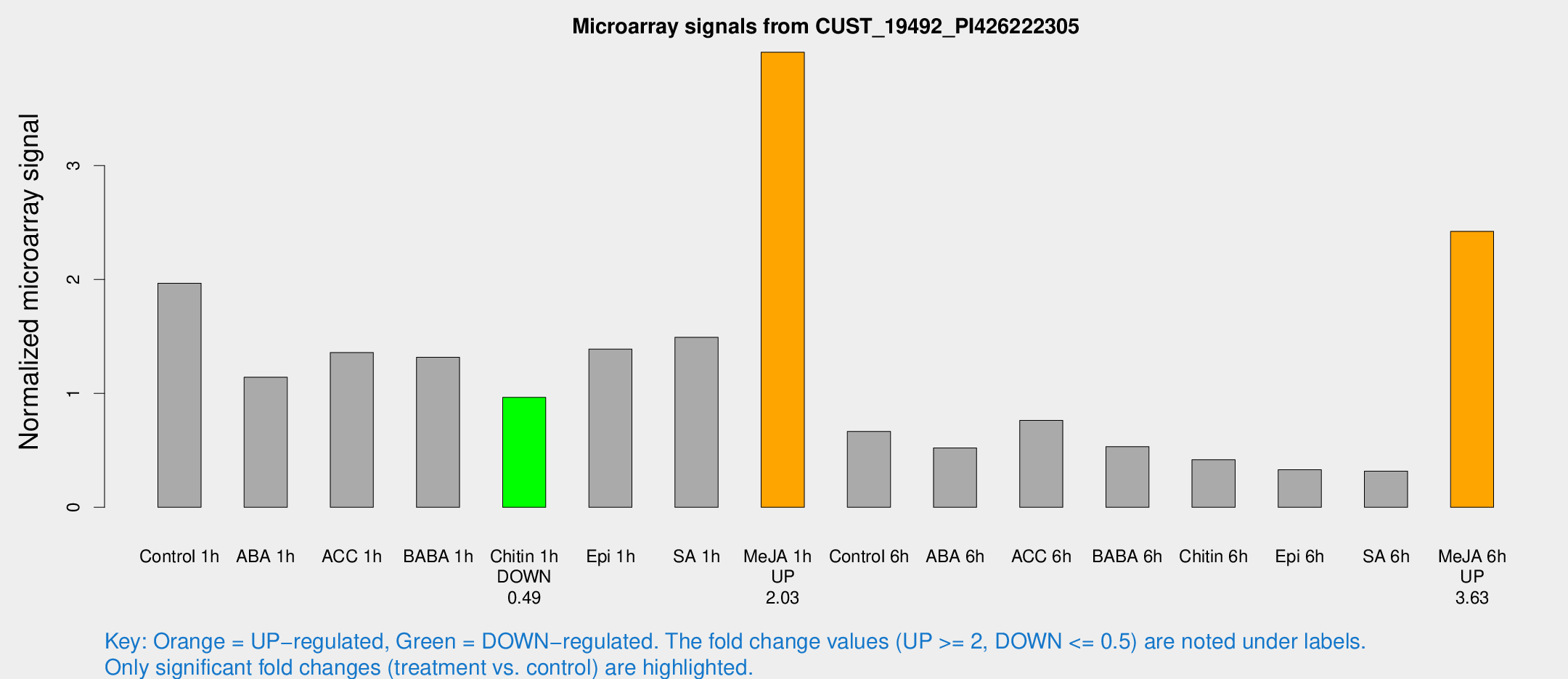
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 260.585 | 65.7613 | 1.96787 | 0.350693 |
| ABA 1h | 130.299 | 24.585 | 1.14178 | 0.135405 |
| ACC 1h | 196.848 | 57.7647 | 1.35837 | 0.43471 |
| BABA 1h | 165.385 | 33.419 | 1.31696 | 0.162718 |
| Chitin 1h | 109.479 | 10.63 | 0.965375 | 0.0672406 |
| Epi 1h | 154.168 | 24.5871 | 1.38954 | 0.197538 |
| SA 1h | 198.942 | 40.4353 | 1.49204 | 0.333549 |
| Me-JA 1h | 413.78 | 51.8197 | 3.99578 | 0.456895 |
| Control 6h | 92.3175 | 25.9305 | 0.66667 | 0.174225 |
| ABA 6h | 69.5437 | 5.29939 | 0.521463 | 0.0441513 |
| ACC 6h | 145.05 | 76.0869 | 0.763181 | 0.363253 |
| BABA 6h | 74.9327 | 5.75149 | 0.533088 | 0.0408422 |
| Chitin 6h | 56.3003 | 6.13514 | 0.41822 | 0.0597543 |
| Epi 6h | 49.2243 | 12.0209 | 0.329939 | 0.0940994 |
| SA 6h | 41.2029 | 8.41356 | 0.317111 | 0.0373592 |
| Me-JA 6h | 304.946 | 34.5113 | 2.4228 | 0.14254 |
Source Transcript PGSC0003DMT400023192 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G03230.1 | +1 | 3e-101 | 314 | 185/414 (45%) | Eukaryotic aspartyl protease family protein | chr1:790110-791414 FORWARD LENGTH=434 |
