Probe CUST_19320_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19320_PI426222305 | JHI_St_60k_v1 | DMT400072727 | GAGGGAAAGTTGGAGATTGGAAGAATTATTTTACTGTCGAGATGAATGATAAACTCAATC |
All Microarray Probes Designed to Gene DMG400028302
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19320_PI426222305 | JHI_St_60k_v1 | DMT400072727 | GAGGGAAAGTTGGAGATTGGAAGAATTATTTTACTGTCGAGATGAATGATAAACTCAATC |
Microarray Signals from CUST_19320_PI426222305
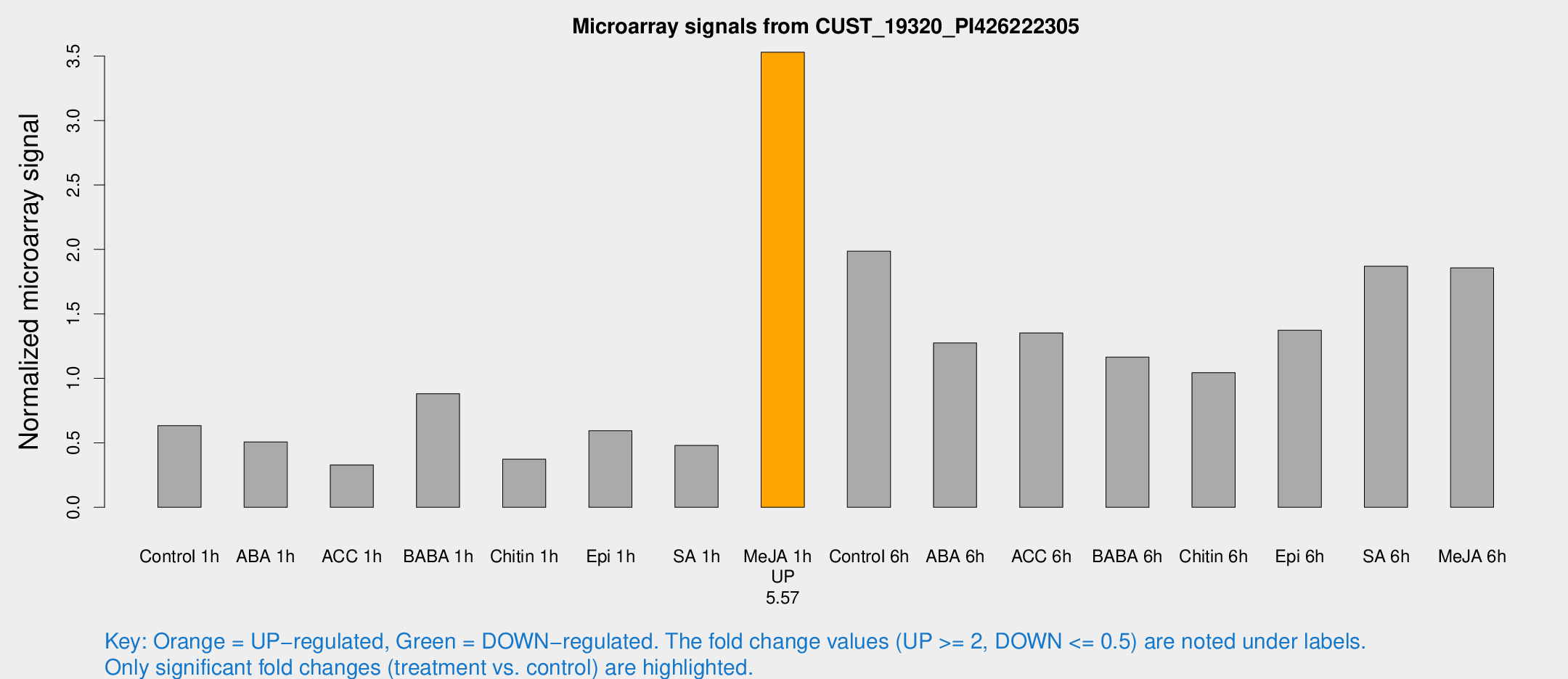
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 26.1571 | 3.53414 | 0.633356 | 0.0865078 |
| ABA 1h | 20.5842 | 5.86029 | 0.507525 | 0.165683 |
| ACC 1h | 14.5251 | 4.07556 | 0.328616 | 0.113371 |
| BABA 1h | 36.317 | 6.60256 | 0.881197 | 0.105817 |
| Chitin 1h | 15.5863 | 4.65944 | 0.372945 | 0.166845 |
| Epi 1h | 23.2976 | 6.23621 | 0.593805 | 0.204275 |
| SA 1h | 22.9928 | 7.71712 | 0.479532 | 0.174159 |
| Me-JA 1h | 122.611 | 23.5898 | 3.52961 | 0.380881 |
| Control 6h | 105.445 | 50.314 | 1.98609 | 1.03682 |
| ABA 6h | 57.4616 | 10.4806 | 1.2746 | 0.157455 |
| ACC 6h | 65.0558 | 8.83378 | 1.35176 | 0.115598 |
| BABA 6h | 59.5569 | 17.5283 | 1.16442 | 0.386132 |
| Chitin 6h | 46.1199 | 4.54214 | 1.04464 | 0.127875 |
| Epi 6h | 65.9204 | 12.1847 | 1.37345 | 0.323185 |
| SA 6h | 79.5218 | 16.6099 | 1.86953 | 0.22856 |
| Me-JA 6h | 76.1922 | 5.51013 | 1.85729 | 0.134258 |
Source Transcript PGSC0003DMT400072727 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G03760.1 | +1 | 1e-116 | 344 | 166/330 (50%) | sulphotransferase 12 | chr2:1149475-1150455 REVERSE LENGTH=326 |
