Probe CUST_19220_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19220_PI426222305 | JHI_St_60k_v1 | DMT400073013 | AATATGTTGTTGTTGTTGTCGTAAGTAGAGTGATGGAATTTGGAAGAGGTGAATTGAAGG |
All Microarray Probes Designed to Gene DMG400028399
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19220_PI426222305 | JHI_St_60k_v1 | DMT400073013 | AATATGTTGTTGTTGTTGTCGTAAGTAGAGTGATGGAATTTGGAAGAGGTGAATTGAAGG |
Microarray Signals from CUST_19220_PI426222305
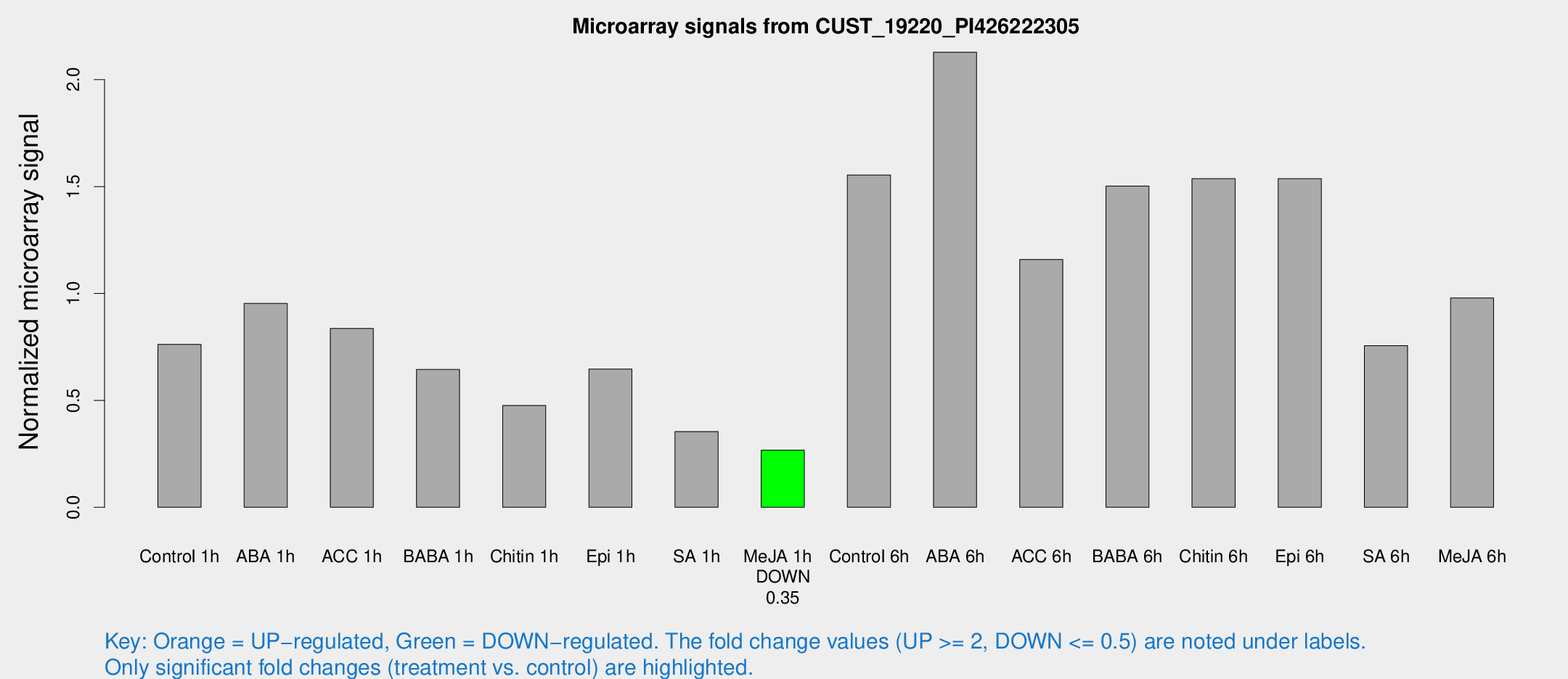
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 56.0773 | 4.77427 | 0.762151 | 0.104123 |
| ABA 1h | 61.8145 | 4.94745 | 0.95375 | 0.0808068 |
| ACC 1h | 65.1659 | 11.8119 | 0.836563 | 0.100251 |
| BABA 1h | 45.4671 | 4.57411 | 0.644618 | 0.0827294 |
| Chitin 1h | 33.4575 | 8.68015 | 0.476305 | 0.117222 |
| Epi 1h | 41.6765 | 5.91753 | 0.646167 | 0.093023 |
| SA 1h | 27.7001 | 5.10208 | 0.354332 | 0.097154 |
| Me-JA 1h | 16.0425 | 3.71242 | 0.267354 | 0.0624217 |
| Control 6h | 121.902 | 29.6725 | 1.55424 | 0.301711 |
| ABA 6h | 166.28 | 12.191 | 2.1281 | 0.132655 |
| ACC 6h | 100.432 | 18.8849 | 1.15908 | 0.0843711 |
| BABA 6h | 126.095 | 19.8781 | 1.50261 | 0.206358 |
| Chitin 6h | 119.76 | 8.00281 | 1.5372 | 0.102739 |
| Epi 6h | 134.101 | 29.1988 | 1.53658 | 0.532357 |
| SA 6h | 65.4794 | 22.1168 | 0.755636 | 0.291986 |
| Me-JA 6h | 77.6608 | 21.7504 | 0.979349 | 0.215969 |
Source Transcript PGSC0003DMT400073013 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
