Probe CUST_19034_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19034_PI426222305 | JHI_St_60k_v1 | DMT400052636 | AAGACCTGATAATTTGTACCTTAAATACAGATGGCAGCCTTTTTCTTGCAACCTGCCAAG |
All Microarray Probes Designed to Gene DMG400020431
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19034_PI426222305 | JHI_St_60k_v1 | DMT400052636 | AAGACCTGATAATTTGTACCTTAAATACAGATGGCAGCCTTTTTCTTGCAACCTGCCAAG |
Microarray Signals from CUST_19034_PI426222305
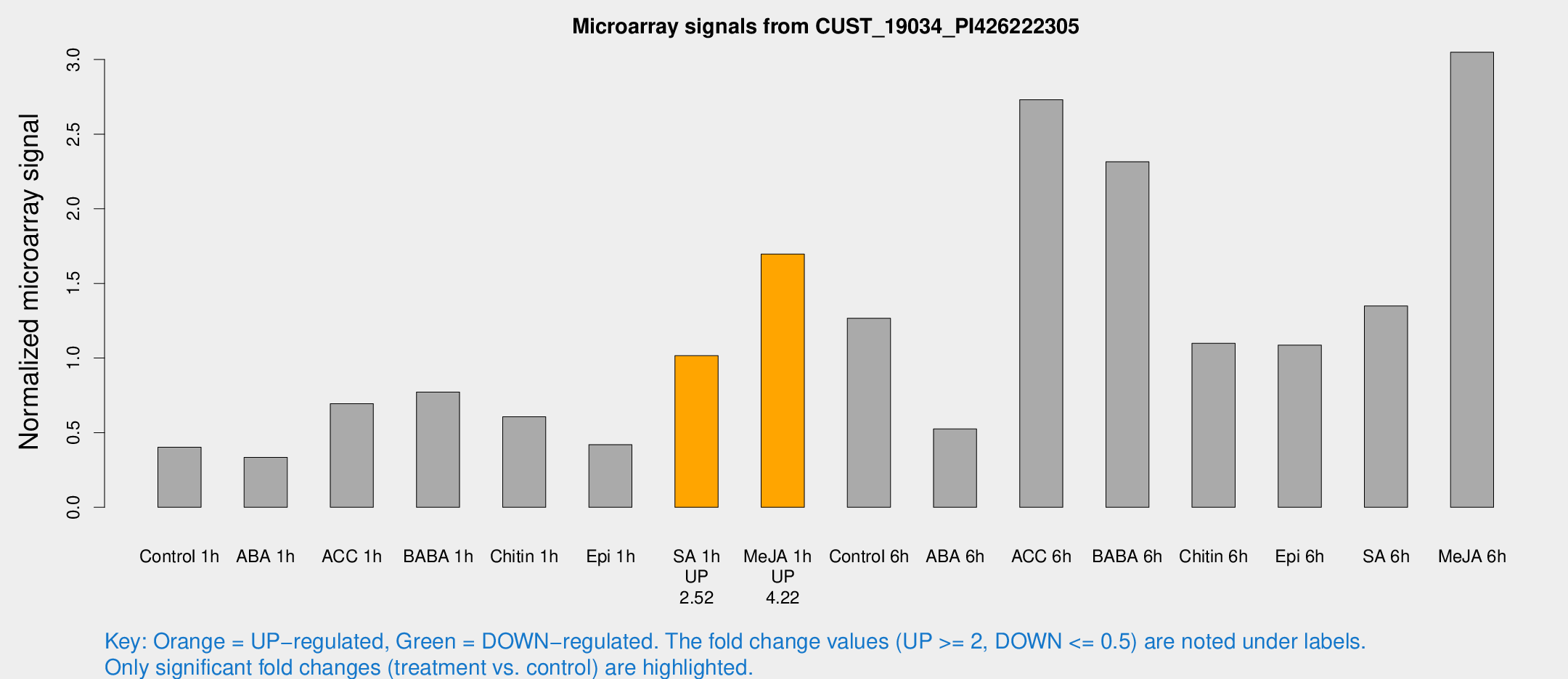
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 58.5819 | 10.4526 | 0.40248 | 0.0478623 |
| ABA 1h | 42.9933 | 7.43187 | 0.334942 | 0.0350568 |
| ACC 1h | 112.455 | 33.6511 | 0.693834 | 0.210265 |
| BABA 1h | 109.216 | 21.6 | 0.772476 | 0.0918595 |
| Chitin 1h | 77.1561 | 5.60832 | 0.606406 | 0.0832923 |
| Epi 1h | 52.0169 | 7.108 | 0.41945 | 0.0439839 |
| SA 1h | 152.926 | 31.9222 | 1.01557 | 0.224078 |
| Me-JA 1h | 197.101 | 20.8201 | 1.69656 | 0.104504 |
| Control 6h | 204.149 | 65.1707 | 1.26689 | 0.394963 |
| ABA 6h | 81.0168 | 13.9529 | 0.524649 | 0.0834889 |
| ACC 6h | 445.388 | 42.8088 | 2.73087 | 0.463209 |
| BABA 6h | 378.833 | 76.2491 | 2.31471 | 0.398767 |
| Chitin 6h | 164.827 | 10.2798 | 1.09891 | 0.068528 |
| Epi 6h | 190.291 | 61.1533 | 1.08634 | 0.393187 |
| SA 6h | 187.91 | 11.4675 | 1.34853 | 0.135414 |
| Me-JA 6h | 431.878 | 44.8945 | 3.04866 | 0.356639 |
Source Transcript PGSC0003DMT400052636 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G42570.1 | +2 | 1e-169 | 488 | 215/317 (68%) | TRICHOME BIREFRINGENCE-LIKE 39 | chr2:17717498-17719921 REVERSE LENGTH=367 |
