Probe CUST_19018_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_19018_PI426222305 | JHI_St_60k_v1 | DMT400044672 | TATATCAGTTTTGCAGATTCAACTCCTTTATTGCCTTCGATCCTATCAGGTCCAAGGTGA |
All Microarray Probes Designed to Gene DMG400017330
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18977_PI426222305 | JHI_St_60k_v1 | DMT400044673 | GCTCGCTGCTGTGTAAATTACTTCTCCAATGTTCACAAAGTTATGATTGTTTTCTTCAAT |
| CUST_19018_PI426222305 | JHI_St_60k_v1 | DMT400044672 | TATATCAGTTTTGCAGATTCAACTCCTTTATTGCCTTCGATCCTATCAGGTCCAAGGTGA |
Microarray Signals from CUST_19018_PI426222305
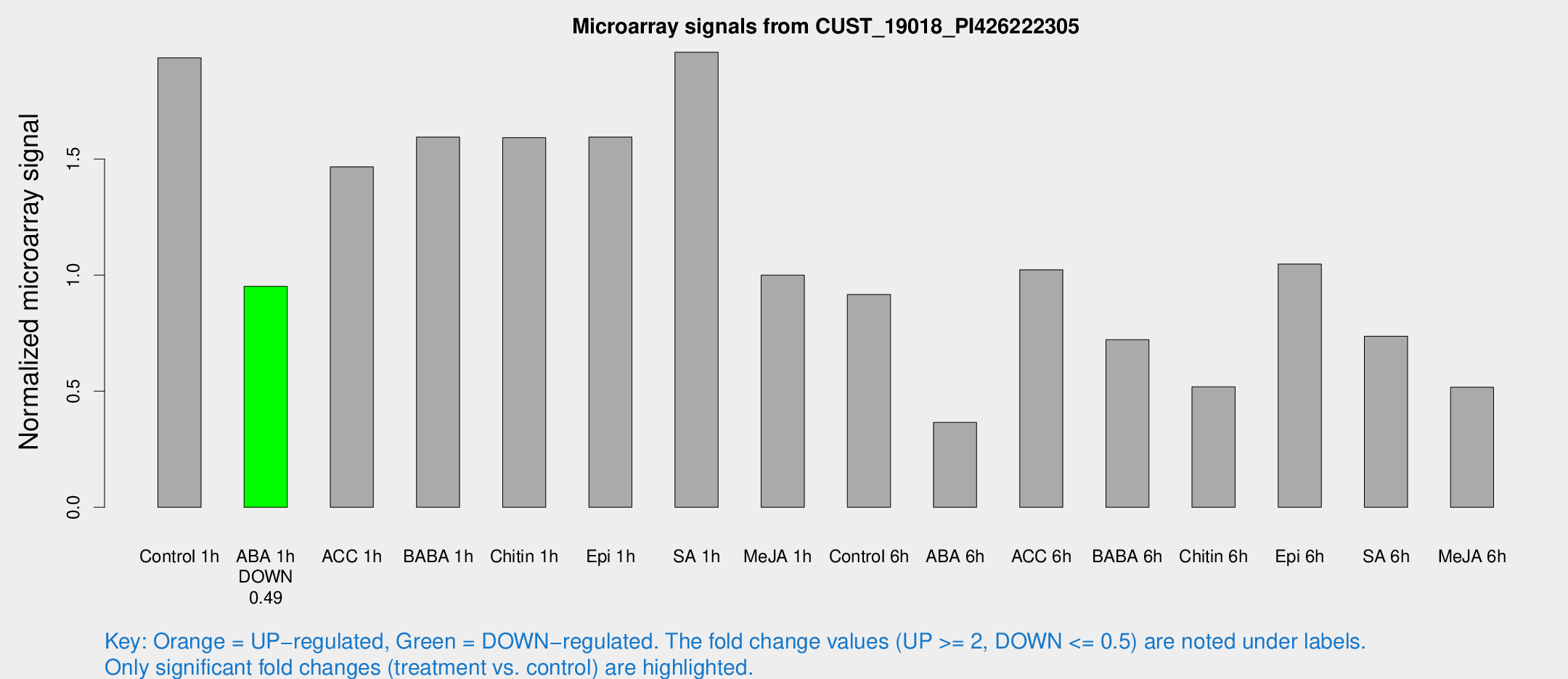
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 489.191 | 123.634 | 1.93632 | 0.345422 |
| ABA 1h | 201.09 | 12.1862 | 0.951371 | 0.112199 |
| ACC 1h | 424.092 | 140.535 | 1.46666 | 0.606211 |
| BABA 1h | 383.69 | 75.8577 | 1.59467 | 0.192588 |
| Chitin 1h | 345.147 | 38.3625 | 1.59196 | 0.0935759 |
| Epi 1h | 330.411 | 22.5736 | 1.59504 | 0.13458 |
| SA 1h | 492.482 | 74.7873 | 1.96027 | 0.217742 |
| Me-JA 1h | 197.598 | 24.8998 | 1.0002 | 0.0610386 |
| Control 6h | 239.211 | 69.3308 | 0.916531 | 0.222522 |
| ABA 6h | 93.8343 | 10.6096 | 0.365754 | 0.0264098 |
| ACC 6h | 286.609 | 42.3745 | 1.023 | 0.161859 |
| BABA 6h | 194.39 | 18.7569 | 0.722331 | 0.0575703 |
| Chitin 6h | 134.019 | 18.6297 | 0.518851 | 0.0577353 |
| Epi 6h | 297.365 | 62.96 | 1.04763 | 0.195514 |
| SA 6h | 180.257 | 36.0613 | 0.736696 | 0.100414 |
| Me-JA 6h | 130.228 | 33.2354 | 0.517019 | 0.100752 |
Source Transcript PGSC0003DMT400044672 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G32300.1 | +1 | 2e-113 | 362 | 236/635 (37%) | S-domain-2 5 | chr4:15599970-15602435 FORWARD LENGTH=821 |
