Probe CUST_18892_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18892_PI426222305 | JHI_St_60k_v1 | DMT400024266 | TGGGGGTTTTTGTGATGATGTTATCACCGGTTTGAGAAATATCTTGATAATAATAGCAAG |
All Microarray Probes Designed to Gene DMG400009378
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18957_PI426222305 | JHI_St_60k_v1 | DMT400024265 | CATCAAGAATTTTGGAGATTATGGGAATTGATTGTCTCACTCGTCATAATATTGCCAGTC |
| CUST_18892_PI426222305 | JHI_St_60k_v1 | DMT400024266 | TGGGGGTTTTTGTGATGATGTTATCACCGGTTTGAGAAATATCTTGATAATAATAGCAAG |
Microarray Signals from CUST_18892_PI426222305
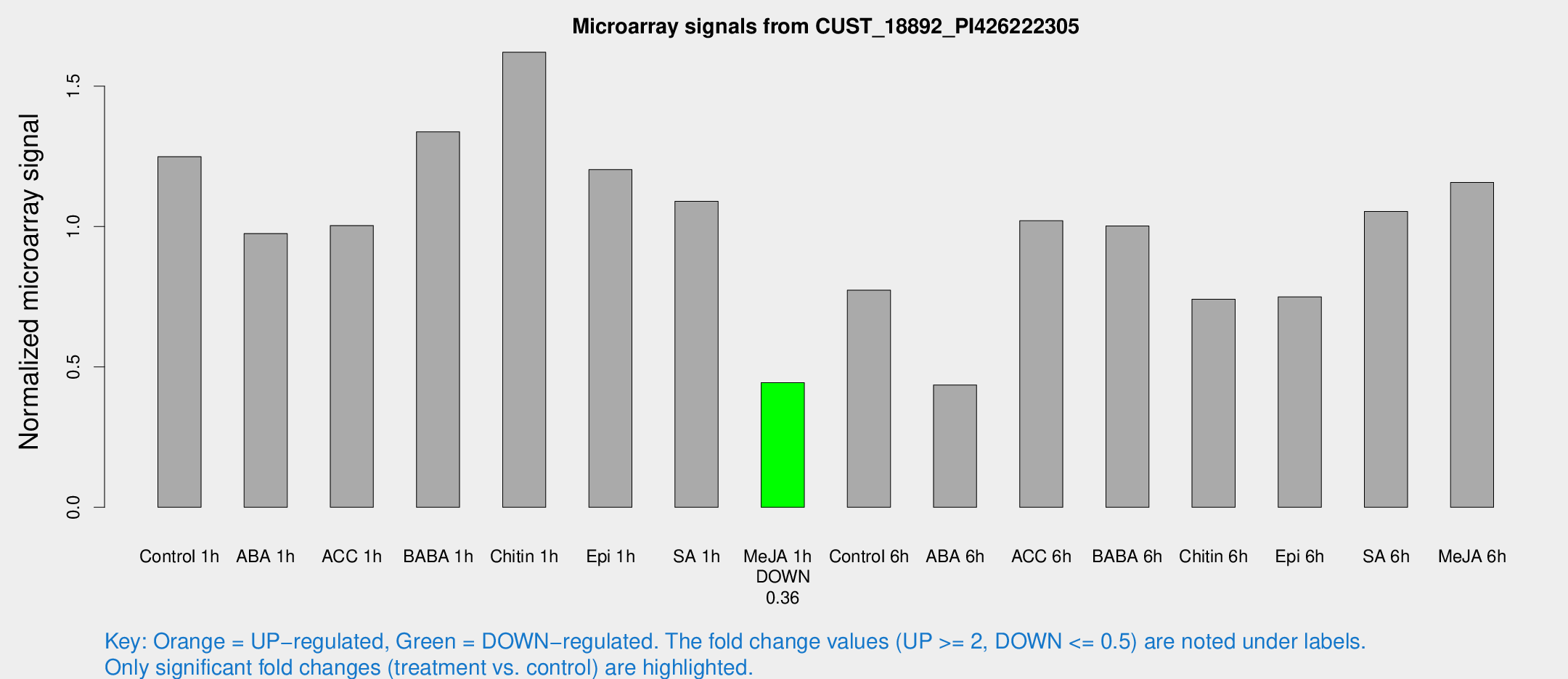
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 1677.72 | 97.1132 | 1.24888 | 0.0721352 |
| ABA 1h | 1192.52 | 211.414 | 0.975237 | 0.138247 |
| ACC 1h | 1392.34 | 119.216 | 1.00369 | 0.125988 |
| BABA 1h | 1811.78 | 357.227 | 1.33767 | 0.162357 |
| Chitin 1h | 1982.55 | 234.205 | 1.621 | 0.0936175 |
| Epi 1h | 1418.45 | 171.802 | 1.20285 | 0.138678 |
| SA 1h | 1566.92 | 315.698 | 1.08975 | 0.219507 |
| Me-JA 1h | 488.976 | 36.3861 | 0.444188 | 0.0257827 |
| Control 6h | 1082.53 | 210.244 | 0.773238 | 0.103107 |
| ABA 6h | 631.674 | 80.2708 | 0.435816 | 0.0519333 |
| ACC 6h | 1588.82 | 168.542 | 1.02078 | 0.0879445 |
| BABA 6h | 1513.98 | 119.884 | 1.00222 | 0.0579057 |
| Chitin 6h | 1065.66 | 95.4966 | 0.740921 | 0.0788354 |
| Epi 6h | 1141.95 | 98.1513 | 0.74961 | 0.0433359 |
| SA 6h | 1414.66 | 154.166 | 1.05386 | 0.0608933 |
| Me-JA 6h | 1675.83 | 422.973 | 1.15719 | 0.26465 |
Source Transcript PGSC0003DMT400024266 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G20570.2 | +3 | 1e-33 | 130 | 116/252 (46%) | GBFs pro-rich region-interacting factor 1 | chr2:8855486-8857522 FORWARD LENGTH=436 |
