Probe CUST_18865_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18865_PI426222305 | JHI_St_60k_v1 | DMT400068224 | AAGAATCTTACAGGAAGATTCTATGCCGGTGATAGGACTGCTAAGGCAAATGGCAAGGAG |
All Microarray Probes Designed to Gene DMG400026524
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18881_PI426222305 | JHI_St_60k_v1 | DMT400068226 | CCTCAAATTCCTCTGAGGAGTTCTCTGAAATATCTTAGTGAAACCATTTATTGATGAAGT |
| CUST_18865_PI426222305 | JHI_St_60k_v1 | DMT400068224 | AAGAATCTTACAGGAAGATTCTATGCCGGTGATAGGACTGCTAAGGCAAATGGCAAGGAG |
Microarray Signals from CUST_18865_PI426222305
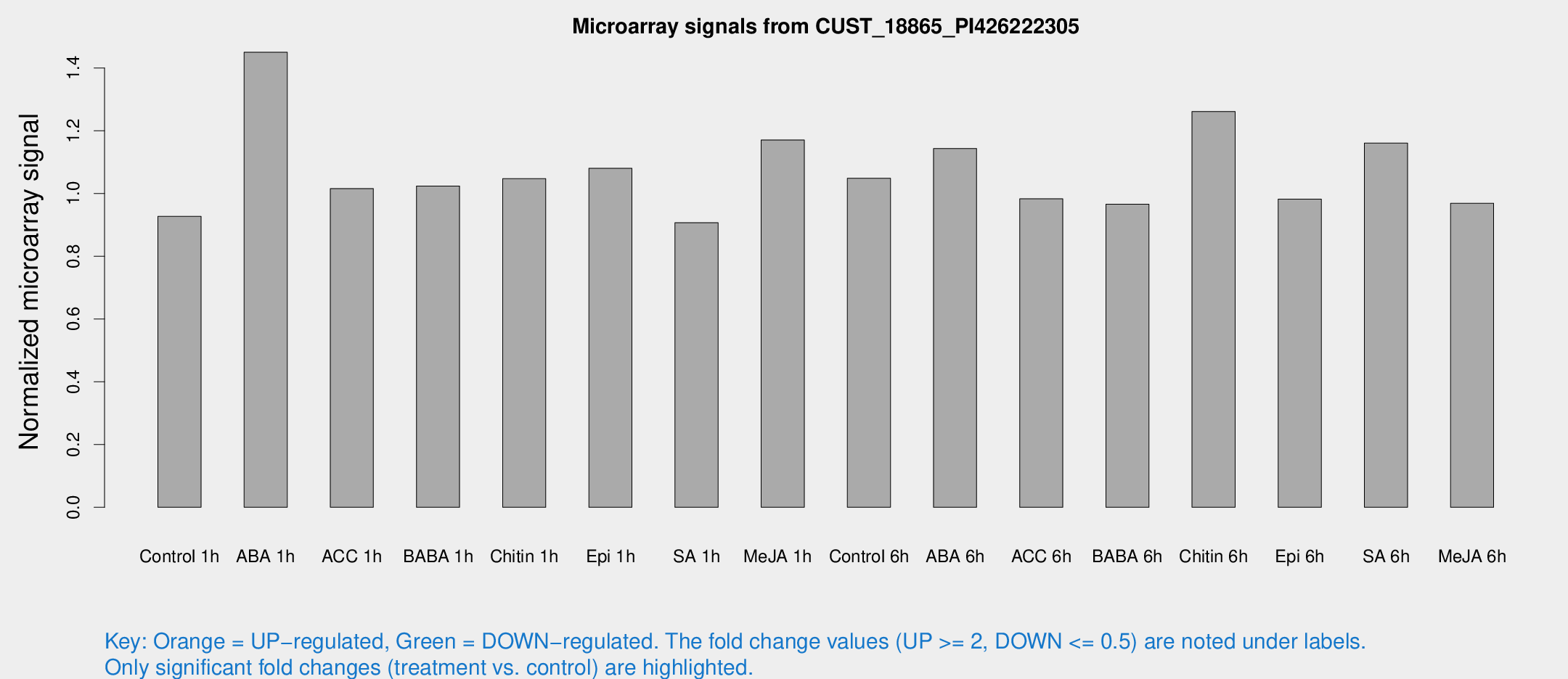
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.72785 | 3.32369 | 0.927153 | 0.537655 |
| ABA 1h | 9.32667 | 3.94812 | 1.45028 | 0.676628 |
| ACC 1h | 6.47236 | 3.65417 | 1.01584 | 0.57403 |
| BABA 1h | 6.09393 | 3.533 | 1.02396 | 0.593564 |
| Chitin 1h | 5.83215 | 3.39581 | 1.04725 | 0.607933 |
| Epi 1h | 5.78373 | 3.35555 | 1.08039 | 0.625624 |
| SA 1h | 5.75414 | 3.33408 | 0.906786 | 0.525108 |
| Me-JA 1h | 5.91083 | 3.42826 | 1.17079 | 0.677905 |
| Control 6h | 6.52048 | 3.49002 | 1.0487 | 0.567256 |
| ABA 6h | 7.81981 | 3.61517 | 1.14354 | 0.571124 |
| ACC 6h | 7.16656 | 4.27698 | 0.983201 | 0.569348 |
| BABA 6h | 6.69199 | 3.88668 | 0.965966 | 0.559614 |
| Chitin 6h | 8.45416 | 3.81203 | 1.26136 | 0.589355 |
| Epi 6h | 6.8834 | 4.01441 | 0.981899 | 0.568575 |
| SA 6h | 7.18375 | 3.63258 | 1.16062 | 0.604029 |
| Me-JA 6h | 5.96088 | 3.45432 | 0.968834 | 0.561013 |
Source Transcript PGSC0003DMT400068224 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G27650.1 | +1 | 1e-102 | 307 | 195/316 (62%) | U2 snRNP auxiliary factor small subunit, putative | chr1:9615152-9616042 FORWARD LENGTH=296 |
