Probe CUST_18856_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18856_PI426222305 | JHI_St_60k_v1 | DMT400001097 | AACATTGGAAACAAATACCAAAGGGTTGTGCTTTGAGTTCCCAATTGGAAAAAGATACAA |
All Microarray Probes Designed to Gene DMG400000415
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18736_PI426222305 | JHI_St_60k_v1 | DMT400001098 | CTTTTCTTGCATTTGGGAGCAAAATCTTAAAGGGGTTTTCTTTTCTAGGAATTTGAGTTC |
| CUST_18856_PI426222305 | JHI_St_60k_v1 | DMT400001097 | AACATTGGAAACAAATACCAAAGGGTTGTGCTTTGAGTTCCCAATTGGAAAAAGATACAA |
Microarray Signals from CUST_18856_PI426222305
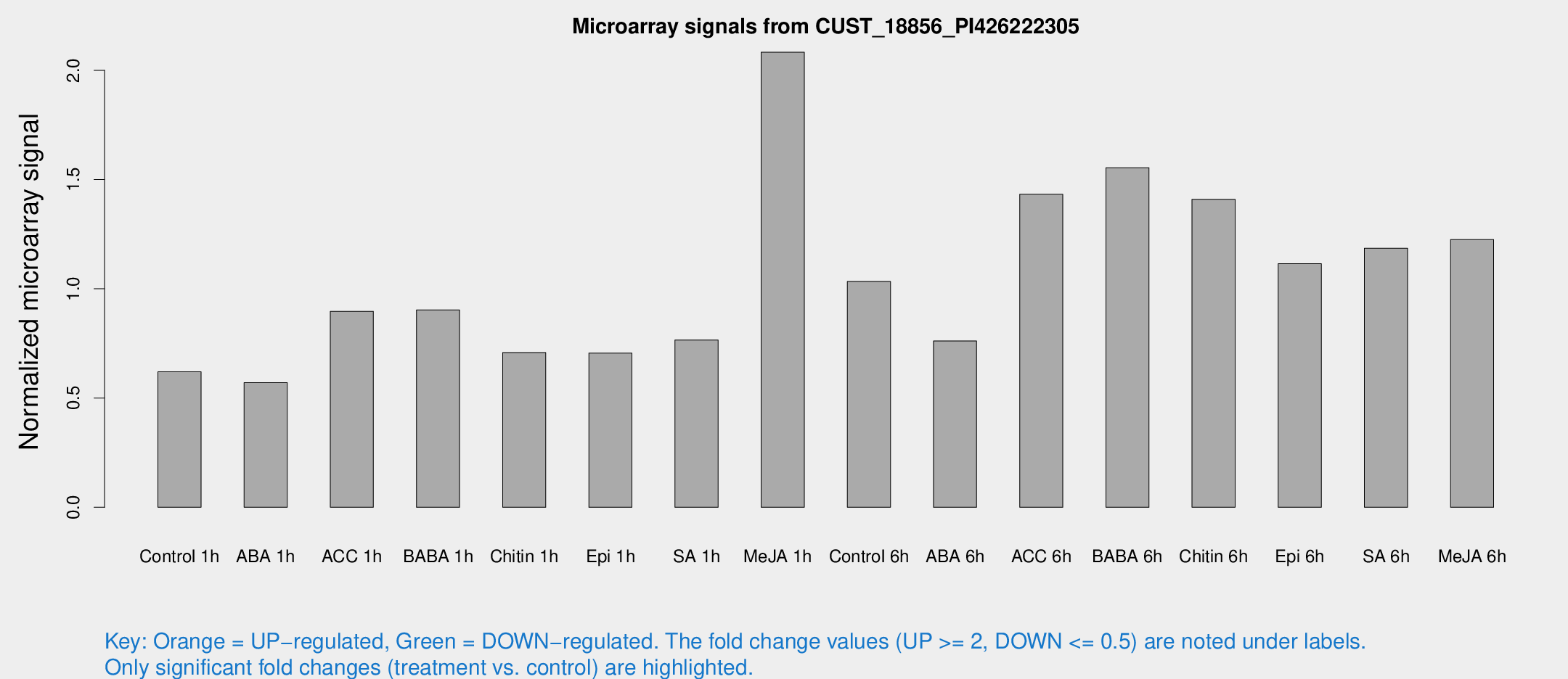
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 205.642 | 43.761 | 0.620243 | 0.0880247 |
| ABA 1h | 168.946 | 35.2925 | 0.570107 | 0.10789 |
| ACC 1h | 352.583 | 129.245 | 0.896686 | 0.400193 |
| BABA 1h | 278.265 | 22.3312 | 0.903028 | 0.0536208 |
| Chitin 1h | 205.445 | 25.7446 | 0.708209 | 0.0427008 |
| Epi 1h | 199.183 | 30.3612 | 0.705991 | 0.106098 |
| SA 1h | 257.615 | 47.0283 | 0.765849 | 0.137616 |
| Me-JA 1h | 559.56 | 110.46 | 2.08273 | 0.239787 |
| Control 6h | 372.322 | 112.862 | 1.03351 | 0.317072 |
| ABA 6h | 263.191 | 39.5196 | 0.761038 | 0.141951 |
| ACC 6h | 526.288 | 48.9595 | 1.43276 | 0.083508 |
| BABA 6h | 566.702 | 91.4964 | 1.5544 | 0.18953 |
| Chitin 6h | 476.965 | 27.8729 | 1.40969 | 0.0823052 |
| Epi 6h | 401.129 | 27.8166 | 1.11517 | 0.112085 |
| SA 6h | 396.232 | 103.866 | 1.18527 | 0.47609 |
| Me-JA 6h | 398.497 | 68.2045 | 1.22509 | 0.10811 |
Source Transcript PGSC0003DMT400001097 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G22930.1 | +2 | 9e-36 | 132 | 107/252 (42%) | Protein of unknown function (DUF1635) | chr5:7668229-7669315 REVERSE LENGTH=238 |
