Probe CUST_18277_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18277_PI426222305 | JHI_St_60k_v1 | DMT400042202 | TTCTATTGTCTCAAATTGTTGGAAGCAACAGGCATCTCTACTGTCCCGGGTTCGGGTTTT |
All Microarray Probes Designed to Gene DMG400016373
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_18277_PI426222305 | JHI_St_60k_v1 | DMT400042202 | TTCTATTGTCTCAAATTGTTGGAAGCAACAGGCATCTCTACTGTCCCGGGTTCGGGTTTT |
Microarray Signals from CUST_18277_PI426222305
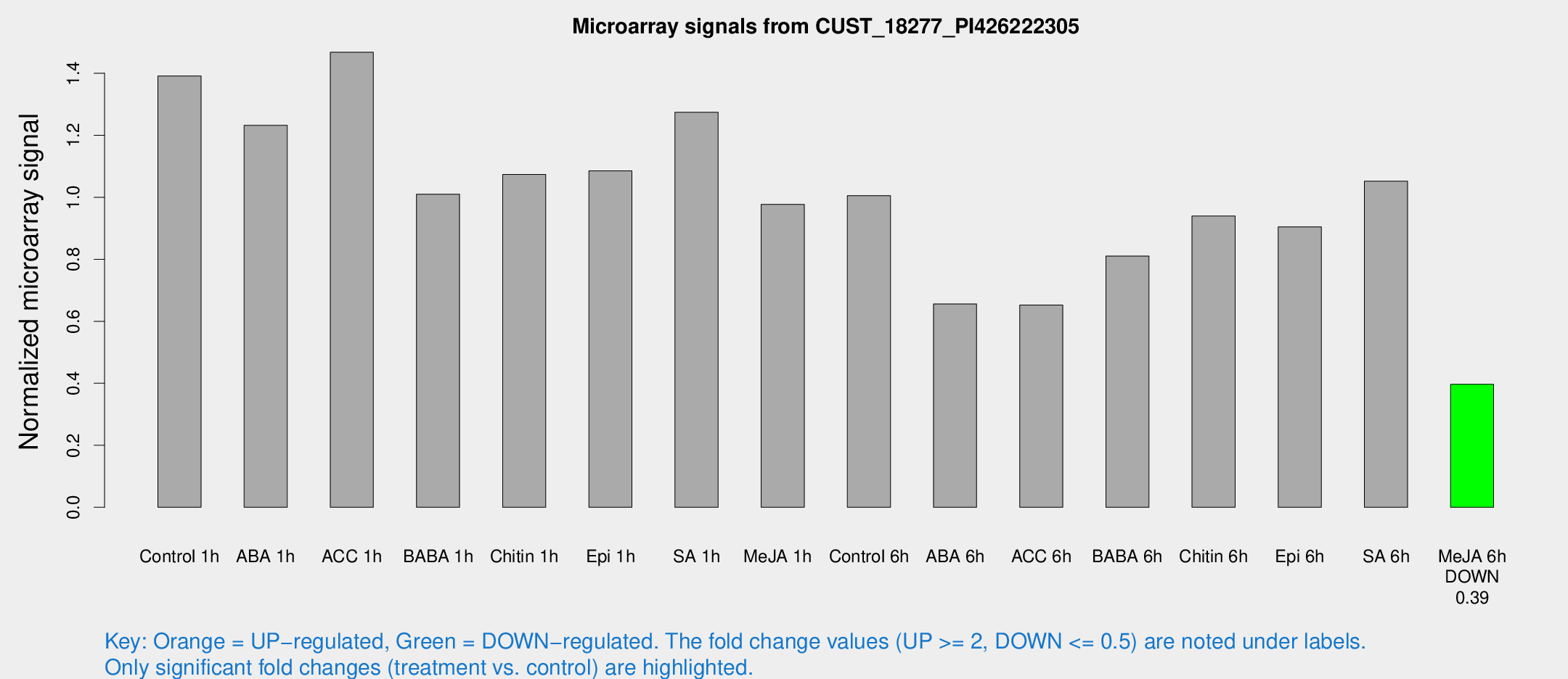
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 14764.1 | 1365.99 | 1.39122 | 0.0822428 |
| ABA 1h | 11561.8 | 1004.67 | 1.23203 | 0.158596 |
| ACC 1h | 16041.9 | 1733.63 | 1.46778 | 0.276211 |
| BABA 1h | 10878.3 | 2389.9 | 1.00998 | 0.154871 |
| Chitin 1h | 10172.6 | 587.861 | 1.07382 | 0.0619977 |
| Epi 1h | 9950.98 | 783.249 | 1.08529 | 0.0817988 |
| SA 1h | 13883.4 | 1195.55 | 1.27417 | 0.0735648 |
| Me-JA 1h | 8428 | 505.119 | 0.977138 | 0.0564163 |
| Control 6h | 10760.8 | 1265.29 | 1.0053 | 0.0592422 |
| ABA 6h | 7342.27 | 424.08 | 0.655868 | 0.0378678 |
| ACC 6h | 8049.17 | 1231.9 | 0.652201 | 0.0376561 |
| BABA 6h | 9752.27 | 1348.49 | 0.810293 | 0.147502 |
| Chitin 6h | 10715.8 | 1379.27 | 0.939431 | 0.100587 |
| Epi 6h | 10797.7 | 795.103 | 0.904616 | 0.0522289 |
| SA 6h | 12136.2 | 4096.98 | 1.05174 | 0.287779 |
| Me-JA 6h | 4322.64 | 818.826 | 0.39693 | 0.045262 |
Source Transcript PGSC0003DMT400042202 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G23310.1 | +1 | 3e-44 | 153 | 73/87 (84%) | glutamate:glyoxylate aminotransferase | chr1:8268720-8271329 REVERSE LENGTH=481 |
