Probe CUST_17844_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17844_PI426222305 | JHI_St_60k_v1 | DMT400066955 | GAACATACCAGCAAGTTACAACAACAACTTCTTCACCAATCTACTTCTCCAATGGGTGAC |
All Microarray Probes Designed to Gene DMG400026027
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17844_PI426222305 | JHI_St_60k_v1 | DMT400066955 | GAACATACCAGCAAGTTACAACAACAACTTCTTCACCAATCTACTTCTCCAATGGGTGAC |
Microarray Signals from CUST_17844_PI426222305
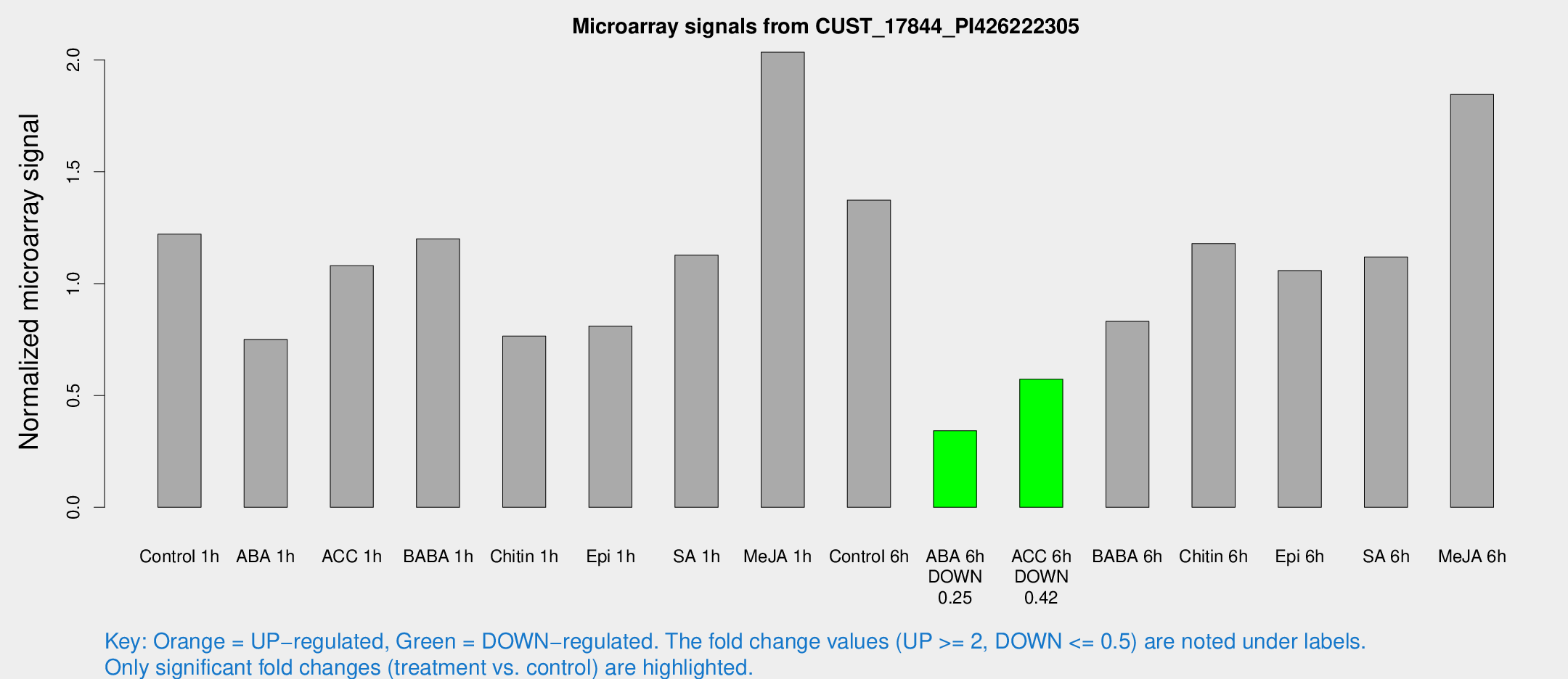
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 193.972 | 35.776 | 1.22119 | 0.224698 |
| ABA 1h | 117.35 | 46.2001 | 0.750378 | 0.256813 |
| ACC 1h | 202.034 | 70.458 | 1.08011 | 0.456162 |
| BABA 1h | 179.014 | 18.6504 | 1.20034 | 0.106781 |
| Chitin 1h | 105.443 | 6.99998 | 0.765733 | 0.0665372 |
| Epi 1h | 110.469 | 17.3736 | 0.810682 | 0.125187 |
| SA 1h | 201.085 | 62.5066 | 1.12762 | 0.548581 |
| Me-JA 1h | 261.416 | 40.8356 | 2.03471 | 0.431636 |
| Control 6h | 227.87 | 56.1557 | 1.37342 | 0.288739 |
| ABA 6h | 58.3075 | 12.9318 | 0.342192 | 0.0903592 |
| ACC 6h | 104.644 | 20.9769 | 0.573048 | 0.0402982 |
| BABA 6h | 148.549 | 31.8393 | 0.83104 | 0.166907 |
| Chitin 6h | 199.933 | 38.7815 | 1.17904 | 0.271186 |
| Epi 6h | 189.441 | 33.533 | 1.05818 | 0.299046 |
| SA 6h | 169.563 | 10.528 | 1.11896 | 0.155102 |
| Me-JA 6h | 291.819 | 55.7371 | 1.84605 | 0.198482 |
Source Transcript PGSC0003DMT400066955 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT1G02065.1 | +2 | 5e-65 | 215 | 140/316 (44%) | squamosa promoter binding protein-like 8 | chr1:365625-367149 FORWARD LENGTH=333 |
