Probe CUST_17718_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17718_PI426222305 | JHI_St_60k_v1 | DMT400067020 | TGCCACCCGTCAGAGTCATCATTGGTAGAATATACCTCTTCAAAGAAGAGAAAATTATAA |
All Microarray Probes Designed to Gene DMG400026049
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17718_PI426222305 | JHI_St_60k_v1 | DMT400067020 | TGCCACCCGTCAGAGTCATCATTGGTAGAATATACCTCTTCAAAGAAGAGAAAATTATAA |
Microarray Signals from CUST_17718_PI426222305
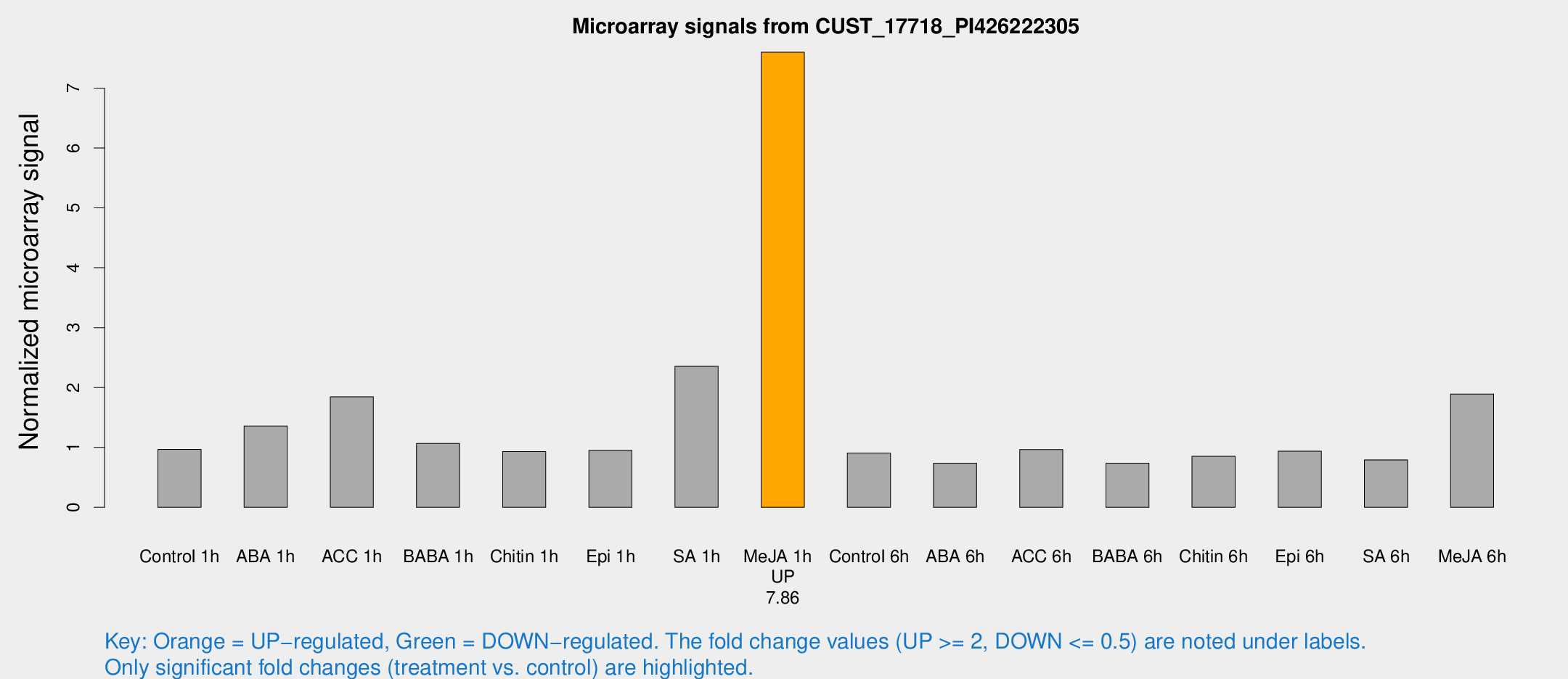
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 9.02819 | 3.66758 | 0.96659 | 0.436437 |
| ABA 1h | 11.6589 | 3.63568 | 1.35784 | 0.517934 |
| ACC 1h | 19.2188 | 6.00305 | 1.84606 | 0.618937 |
| BABA 1h | 9.9486 | 3.97332 | 1.0665 | 0.50881 |
| Chitin 1h | 7.47784 | 3.77371 | 0.93181 | 0.472889 |
| Epi 1h | 7.44112 | 3.67704 | 0.950097 | 0.488195 |
| SA 1h | 31.0494 | 18.0547 | 2.35506 | 1.66405 |
| Me-JA 1h | 55.9857 | 7.36496 | 7.59843 | 1.24653 |
| Control 6h | 8.34406 | 3.91547 | 0.907523 | 0.45571 |
| ABA 6h | 6.95681 | 4.04137 | 0.735614 | 0.426589 |
| ACC 6h | 10.3175 | 4.8001 | 0.965292 | 0.454576 |
| BABA 6h | 7.34391 | 4.26971 | 0.735913 | 0.426153 |
| Chitin 6h | 8.16388 | 4.20859 | 0.851818 | 0.450412 |
| Epi 6h | 9.53901 | 4.55676 | 0.938058 | 0.467052 |
| SA 6h | 6.9601 | 4.03979 | 0.792305 | 0.45983 |
| Me-JA 6h | 17.1089 | 3.92111 | 1.89194 | 0.465594 |
Source Transcript PGSC0003DMT400067020 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G44840.1 | +1 | 1e-42 | 145 | 72/107 (67%) | ethylene-responsive element binding factor 13 | chr2:18495440-18496120 FORWARD LENGTH=226 |
