Probe CUST_17656_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17656_PI426222305 | JHI_St_60k_v1 | DMT400066809 | GTCATTTTTGTCTGTATACTTTTGACTTTGCCAGCTAAACACATAAACTTGTCCCAAATC |
All Microarray Probes Designed to Gene DMG400025976
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17681_PI426222305 | JHI_St_60k_v1 | DMT400066807 | CACTTCAGAAGTTGGTTCCAAATTCGAGCAAGACAGATAAAGCATCAATGCTAGACGAAG |
| CUST_17765_PI426222305 | JHI_St_60k_v1 | DMT400066808 | CACTTCAGAAGTTGGTTCCAAATTCGAGCAAGACAGATAAAGCATCAATGCTAGACGAAG |
| CUST_17656_PI426222305 | JHI_St_60k_v1 | DMT400066809 | GTCATTTTTGTCTGTATACTTTTGACTTTGCCAGCTAAACACATAAACTTGTCCCAAATC |
Microarray Signals from CUST_17656_PI426222305
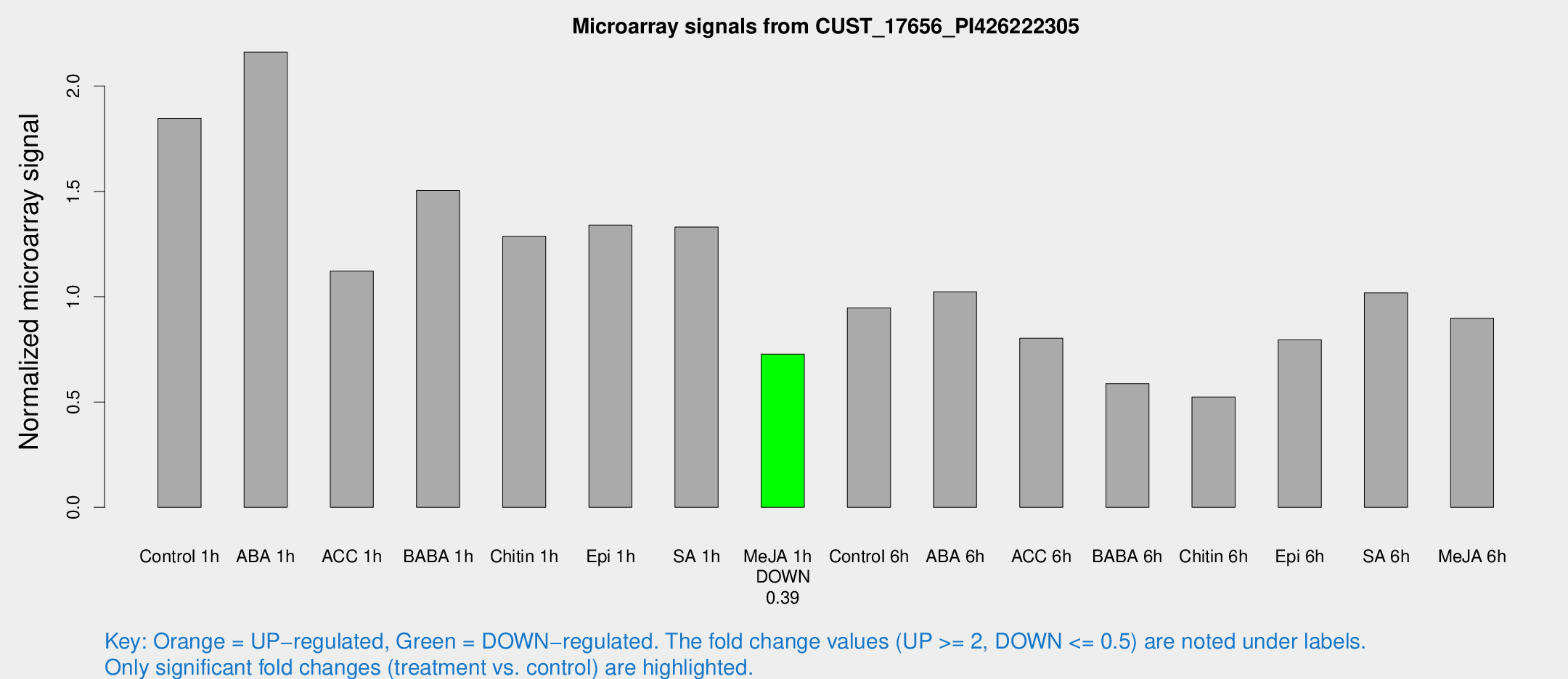
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 6698.87 | 792.87 | 1.84621 | 0.11984 |
| ABA 1h | 6923.93 | 716.934 | 2.16097 | 0.124769 |
| ACC 1h | 4317.88 | 860.325 | 1.12131 | 0.170117 |
| BABA 1h | 5502.72 | 1187.77 | 1.50433 | 0.224032 |
| Chitin 1h | 4199.72 | 499.48 | 1.28688 | 0.0937865 |
| Epi 1h | 4279.08 | 683.381 | 1.34062 | 0.21897 |
| SA 1h | 5048.2 | 906.287 | 1.33039 | 0.194482 |
| Me-JA 1h | 2145.48 | 208.691 | 0.727082 | 0.0586745 |
| Control 6h | 3468.94 | 505.156 | 0.946707 | 0.098317 |
| ABA 6h | 3939.56 | 392.661 | 1.02368 | 0.0680202 |
| ACC 6h | 3354.94 | 416.882 | 0.802879 | 0.0825071 |
| BABA 6h | 2363.37 | 136.92 | 0.587757 | 0.0339499 |
| Chitin 6h | 2025.26 | 237.201 | 0.524302 | 0.0435783 |
| Epi 6h | 3309.65 | 537.261 | 0.795406 | 0.0851644 |
| SA 6h | 3673.85 | 491.849 | 1.01842 | 0.0622297 |
| Me-JA 6h | 3222.05 | 249.471 | 0.897843 | 0.0518479 |
Source Transcript PGSC0003DMT400066809 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | None | - | - | - | - | - |
