Probe CUST_17495_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17495_PI426222305 | JHI_St_60k_v1 | DMT400008363 | TCTGAGATGTTCATGTACCCAGAAGTGTGGATAAAGAAGATGAACAAGTTGGAATAATAT |
All Microarray Probes Designed to Gene DMG400003225
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17495_PI426222305 | JHI_St_60k_v1 | DMT400008363 | TCTGAGATGTTCATGTACCCAGAAGTGTGGATAAAGAAGATGAACAAGTTGGAATAATAT |
Microarray Signals from CUST_17495_PI426222305
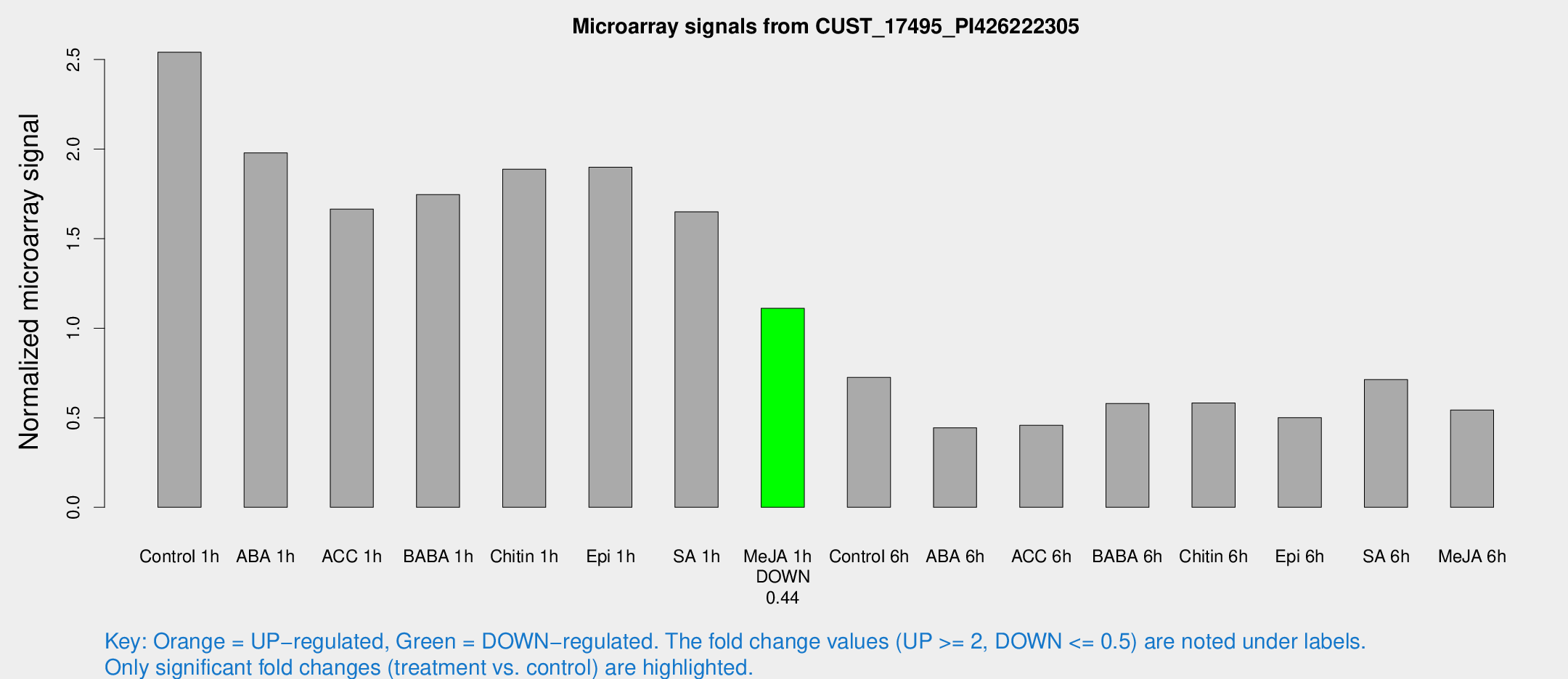
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 3521 | 366.714 | 2.54085 | 0.14672 |
| ABA 1h | 2408.62 | 150.031 | 1.97927 | 0.11431 |
| ACC 1h | 2422.8 | 412.286 | 1.6654 | 0.190657 |
| BABA 1h | 2467.17 | 568.665 | 1.74633 | 0.298912 |
| Chitin 1h | 2355.34 | 253.481 | 1.88784 | 0.109036 |
| Epi 1h | 2260.12 | 138.11 | 1.89841 | 0.109648 |
| SA 1h | 2376.63 | 343.122 | 1.64972 | 0.164993 |
| Me-JA 1h | 1273.83 | 201.048 | 1.11108 | 0.0778872 |
| Control 6h | 1040.92 | 224.647 | 0.72507 | 0.121044 |
| ABA 6h | 654.814 | 68.2125 | 0.444316 | 0.0582913 |
| ACC 6h | 731.95 | 90.2412 | 0.45794 | 0.0610268 |
| BABA 6h | 894.29 | 68.5579 | 0.579802 | 0.0449542 |
| Chitin 6h | 851.938 | 49.5341 | 0.582288 | 0.0337491 |
| Epi 6h | 775.45 | 45.1439 | 0.500237 | 0.0290257 |
| SA 6h | 1019.58 | 231.601 | 0.713471 | 0.100049 |
| Me-JA 6h | 763.92 | 122.865 | 0.543567 | 0.0536022 |
Source Transcript PGSC0003DMT400008363 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00880.1 | +3 | 5e-33 | 119 | 56/72 (78%) | SAUR-like auxin-responsive protein family | chr4:366692-367060 REVERSE LENGTH=122 |
