Probe CUST_17053_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17053_PI426222305 | JHI_St_60k_v1 | DMT400094724 | TTGTTAACACTGTTCCAGCGACTGAACTAAACCAAAGCAGTGGCTCAAAGGCAGTTGATG |
All Microarray Probes Designed to Gene DMG400044295
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17053_PI426222305 | JHI_St_60k_v1 | DMT400094724 | TTGTTAACACTGTTCCAGCGACTGAACTAAACCAAAGCAGTGGCTCAAAGGCAGTTGATG |
Microarray Signals from CUST_17053_PI426222305
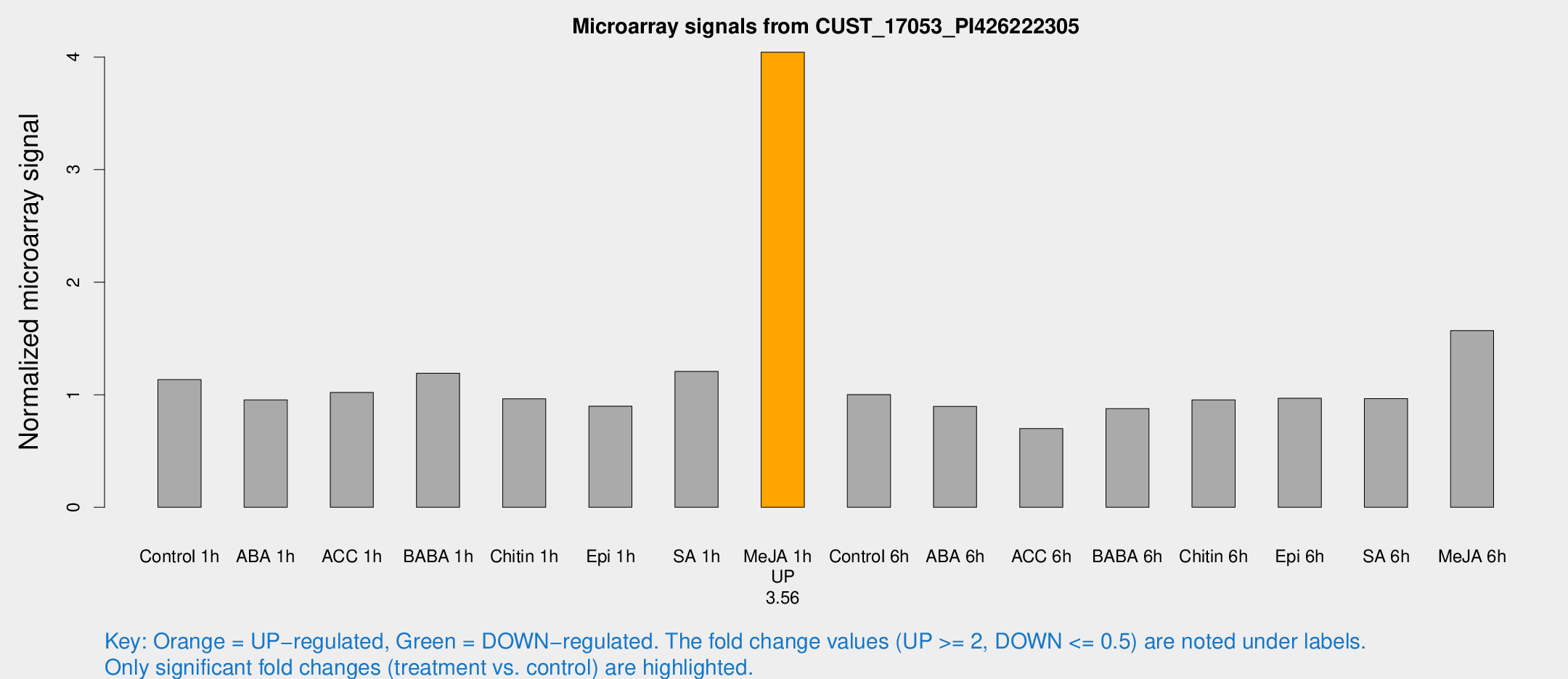
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 486.484 | 82.4801 | 1.13429 | 0.180535 |
| ABA 1h | 373.881 | 96.9118 | 0.954801 | 0.172361 |
| ACC 1h | 469.947 | 127.406 | 1.01966 | 0.231372 |
| BABA 1h | 480.334 | 33.4553 | 1.19124 | 0.0693738 |
| Chitin 1h | 362.636 | 25.1789 | 0.963994 | 0.113158 |
| Epi 1h | 337.473 | 66.2645 | 0.898553 | 0.18677 |
| SA 1h | 533.748 | 99.6491 | 1.20718 | 0.260158 |
| Me-JA 1h | 1374.53 | 79.4806 | 4.0427 | 0.280439 |
| Control 6h | 448.949 | 107.282 | 1.00112 | 0.197987 |
| ABA 6h | 397.44 | 23.286 | 0.896824 | 0.0524709 |
| ACC 6h | 338.169 | 39.5984 | 0.69907 | 0.0413025 |
| BABA 6h | 411.736 | 37.4803 | 0.876697 | 0.0826711 |
| Chitin 6h | 432.414 | 66.1102 | 0.954895 | 0.141809 |
| Epi 6h | 462.644 | 61.0698 | 0.968567 | 0.176692 |
| SA 6h | 400.688 | 35.3701 | 0.965306 | 0.05654 |
| Me-JA 6h | 653.895 | 44.68 | 1.56985 | 0.0910849 |
Source Transcript PGSC0003DMT400094724 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT4G00650.1 | +1 | 7e-29 | 119 | 72/171 (42%) | FRIGIDA-like protein | chr4:269026-270363 FORWARD LENGTH=314 |
