Probe CUST_17019_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17019_PI426222305 | JHI_St_60k_v1 | DMT400018461 | GAGATGGAGAGGATTTTGGCTCTAATATATATGGAGTTTGCGCGTGATTGCTTAGCTTAG |
All Microarray Probes Designed to Gene DMG400007162
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_17019_PI426222305 | JHI_St_60k_v1 | DMT400018461 | GAGATGGAGAGGATTTTGGCTCTAATATATATGGAGTTTGCGCGTGATTGCTTAGCTTAG |
Microarray Signals from CUST_17019_PI426222305
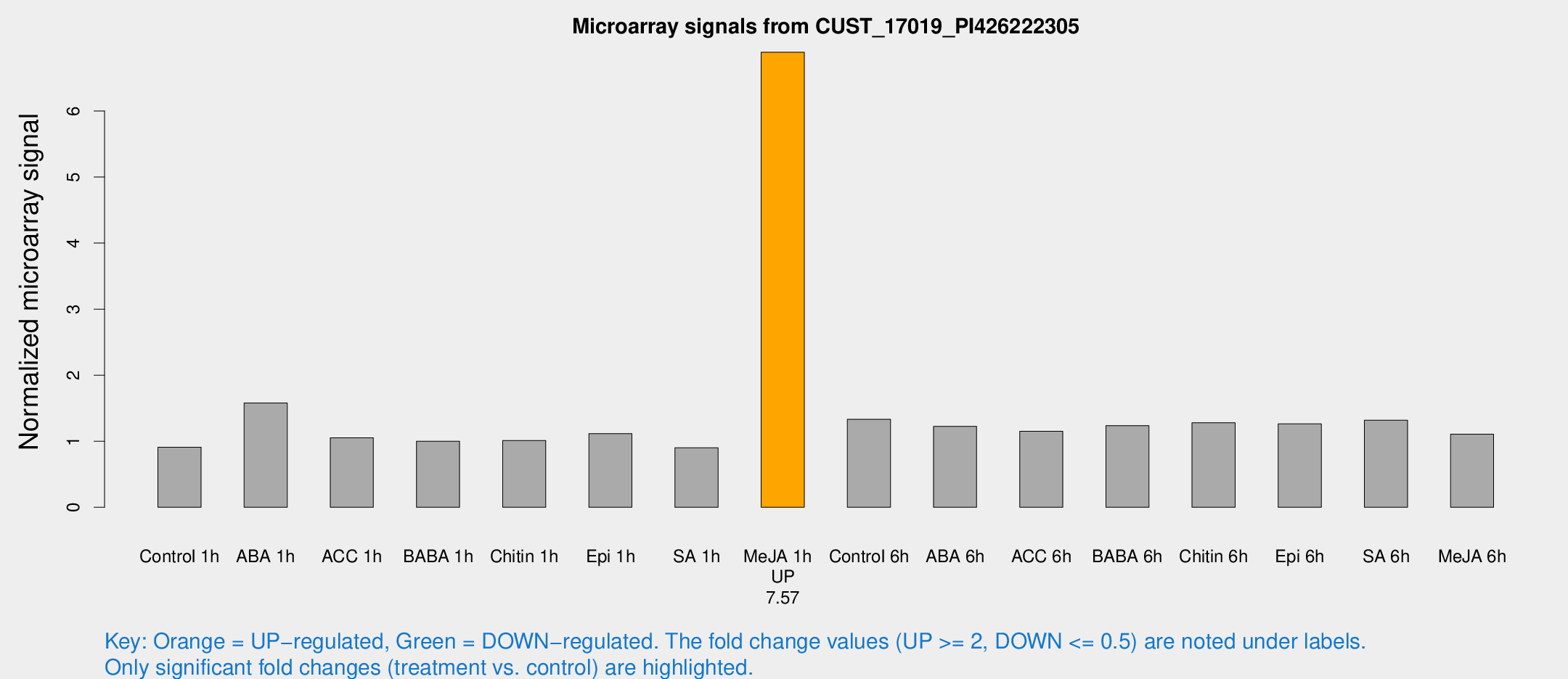
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 5.50455 | 3.19389 | 0.910627 | 0.527712 |
| ABA 1h | 9.12155 | 3.14455 | 1.5788 | 0.639094 |
| ACC 1h | 6.65804 | 3.93956 | 1.05211 | 0.609262 |
| BABA 1h | 5.82876 | 3.37949 | 1.00043 | 0.579337 |
| Chitin 1h | 5.48207 | 3.18809 | 1.01135 | 0.58375 |
| Epi 1h | 5.9071 | 3.15103 | 1.11637 | 0.604533 |
| SA 1h | 5.58616 | 3.23844 | 0.900313 | 0.521674 |
| Me-JA 1h | 33.9834 | 3.85009 | 6.88934 | 0.781436 |
| Control 6h | 9.2021 | 3.53447 | 1.3312 | 0.621949 |
| ABA 6h | 8.52501 | 3.4047 | 1.22575 | 0.582657 |
| ACC 6h | 8.15892 | 4.08347 | 1.15114 | 0.580099 |
| BABA 6h | 9.35438 | 3.69948 | 1.23647 | 0.592392 |
| Chitin 6h | 8.83433 | 3.65364 | 1.2805 | 0.615045 |
| Epi 6h | 9.22314 | 3.9015 | 1.2637 | 0.60905 |
| SA 6h | 8.0366 | 3.50056 | 1.3177 | 0.602774 |
| Me-JA 6h | 6.90475 | 3.30266 | 1.1076 | 0.5633 |
Source Transcript PGSC0003DMT400018461 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT2G28085.1 | +1 | 1e-23 | 91 | 47/76 (62%) | SAUR-like auxin-responsive protein family | chr2:11968182-11968556 REVERSE LENGTH=124 |
