Probe CUST_16821_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16821_PI426222305 | JHI_St_60k_v1 | DMT400069641 | GCAATTCGTTGTTCTGCGCCTGAATGCTGATGTTTGTTTTTCTTTCGAAAATAATATTTC |
All Microarray Probes Designed to Gene DMG400027075
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16821_PI426222305 | JHI_St_60k_v1 | DMT400069641 | GCAATTCGTTGTTCTGCGCCTGAATGCTGATGTTTGTTTTTCTTTCGAAAATAATATTTC |
Microarray Signals from CUST_16821_PI426222305
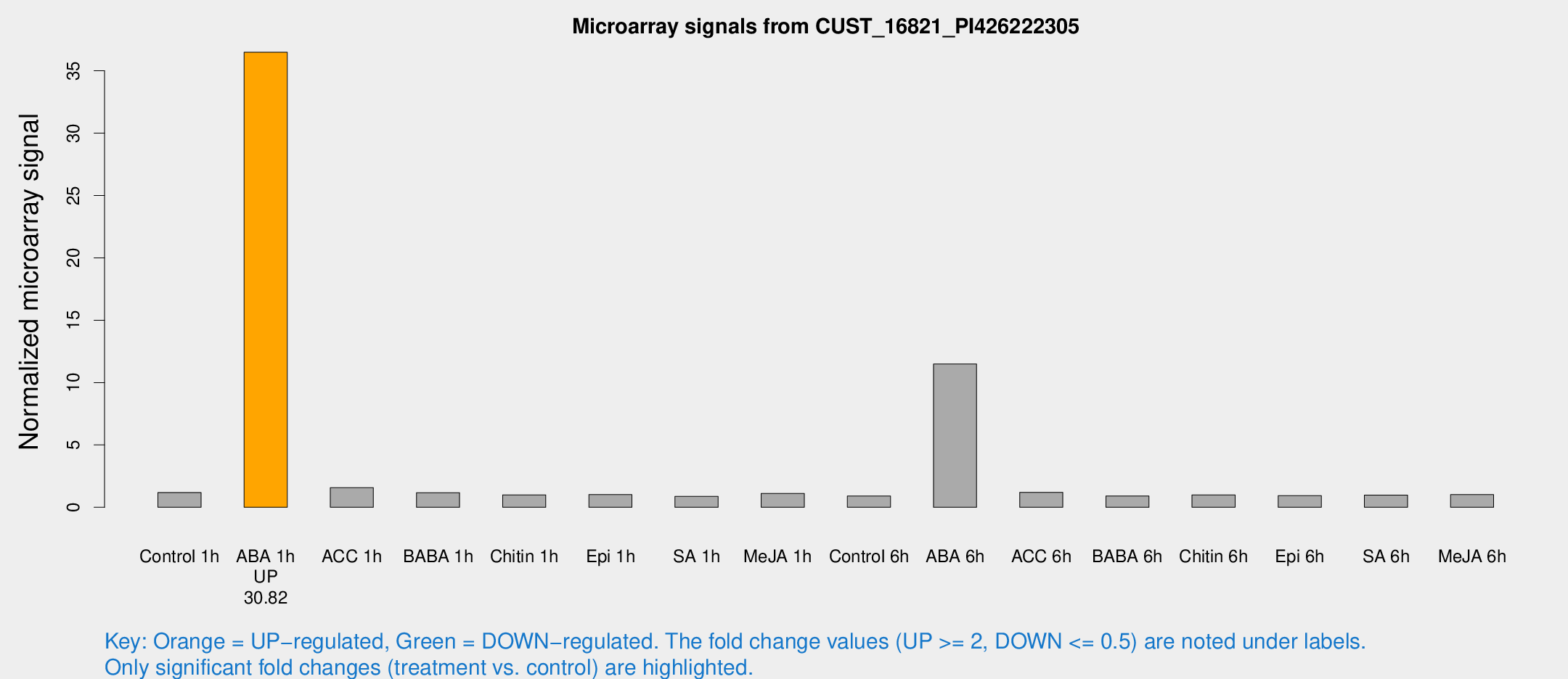
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 7.65482 | 3.06403 | 1.18404 | 0.562632 |
| ABA 1h | 193.465 | 20.6205 | 36.4897 | 3.26839 |
| ACC 1h | 10.2408 | 3.68234 | 1.56985 | 0.661838 |
| BABA 1h | 6.89252 | 3.23401 | 1.17188 | 0.586587 |
| Chitin 1h | 5.21994 | 3.03493 | 0.997477 | 0.565853 |
| Epi 1h | 5.15434 | 2.99119 | 1.0211 | 0.581618 |
| SA 1h | 5.31743 | 3.08216 | 0.873519 | 0.506081 |
| Me-JA 1h | 5.40497 | 3.13931 | 1.11437 | 0.645235 |
| Control 6h | 5.35405 | 3.10115 | 0.901791 | 0.522283 |
| ABA 6h | 79.114 | 20.4378 | 11.4927 | 3.31967 |
| ACC 6h | 8.55377 | 3.86327 | 1.20218 | 0.582794 |
| BABA 6h | 6.03226 | 3.5038 | 0.90813 | 0.526451 |
| Chitin 6h | 6.29105 | 3.47323 | 0.996392 | 0.551975 |
| Epi 6h | 6.28406 | 3.66735 | 0.934117 | 0.541058 |
| SA 6h | 5.7282 | 3.31964 | 0.976921 | 0.565717 |
| Me-JA 6h | 6.06913 | 3.13841 | 1.01578 | 0.537152 |
Source Transcript PGSC0003DMT400069641 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G63130.1 | +3 | 5e-33 | 123 | 62/110 (56%) | Octicosapeptide/Phox/Bem1p family protein | chr5:25323190-25323857 FORWARD LENGTH=192 |
