Probe CUST_16806_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16806_PI426222305 | JHI_St_60k_v1 | DMT400069254 | GTTGTCTCCTGTTTCCTACAAACATGACTTTTTCAATTAGAAAGACCTTTGTATTGTGAC |
All Microarray Probes Designed to Gene DMG402026939
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16806_PI426222305 | JHI_St_60k_v1 | DMT400069254 | GTTGTCTCCTGTTTCCTACAAACATGACTTTTTCAATTAGAAAGACCTTTGTATTGTGAC |
Microarray Signals from CUST_16806_PI426222305
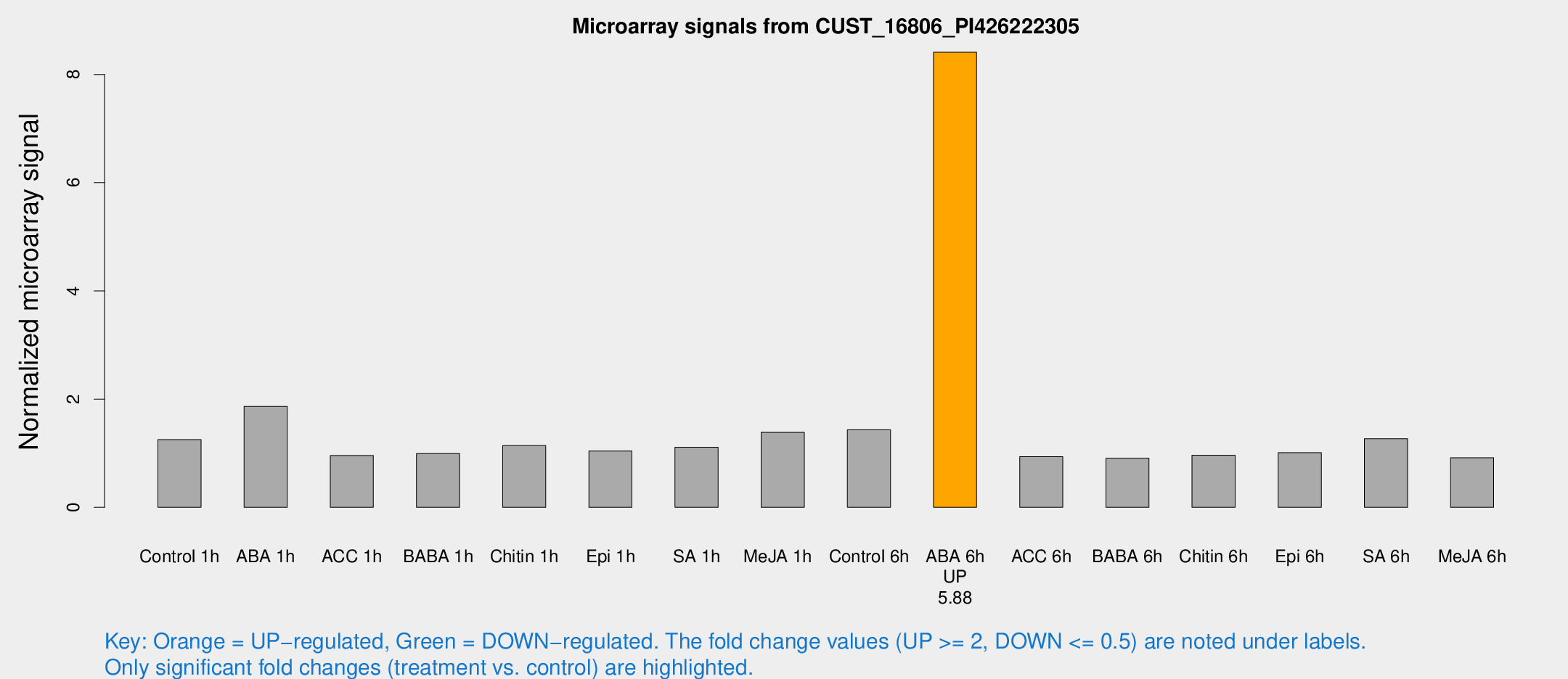
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 8.69172 | 3.23364 | 1.24995 | 0.589201 |
| ABA 1h | 11.3061 | 3.36563 | 1.86623 | 0.670831 |
| ACC 1h | 6.04526 | 3.5078 | 0.957733 | 0.555071 |
| BABA 1h | 5.88736 | 3.41858 | 0.993756 | 0.576397 |
| Chitin 1h | 6.39985 | 3.18888 | 1.13912 | 0.581459 |
| Epi 1h | 5.53031 | 3.1538 | 1.04002 | 0.591115 |
| SA 1h | 7.42512 | 3.16906 | 1.10889 | 0.538109 |
| Me-JA 1h | 7.11427 | 3.26962 | 1.3866 | 0.662353 |
| Control 6h | 10.8198 | 5.12568 | 1.43094 | 0.659028 |
| ABA 6h | 55.8288 | 7.51007 | 8.4107 | 0.723735 |
| ACC 6h | 6.75679 | 4.01094 | 0.937748 | 0.543329 |
| BABA 6h | 6.25941 | 3.63196 | 0.909334 | 0.526692 |
| Chitin 6h | 6.30281 | 3.65219 | 0.96398 | 0.558268 |
| Epi 6h | 7.09459 | 3.7829 | 1.01058 | 0.540368 |
| SA 6h | 7.91467 | 3.47432 | 1.26694 | 0.588867 |
| Me-JA 6h | 5.60169 | 3.24543 | 0.916038 | 0.530551 |
Source Transcript PGSC0003DMT400069254 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT5G50700.1 | +1 | 3e-146 | 423 | 220/349 (63%) | hydroxysteroid dehydrogenase 1 | chr5:20623259-20624995 REVERSE LENGTH=349 |
