Probe CUST_16796_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16796_PI426222305 | JHI_St_60k_v1 | DMT400069540 | TTTGACGTTCTTGAAGTCATTGGCTGCCGAGGGCTGAATCAAACTAACAACATCTGCTTC |
All Microarray Probes Designed to Gene DMG402027033
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16784_PI426222305 | JHI_St_60k_v1 | DMT400069541 | TTTGACGTTCTTGAAGTCATTGGCTGCCGAGGGCTGAATCAAACTAACAACATCTGCTTC |
| CUST_16796_PI426222305 | JHI_St_60k_v1 | DMT400069540 | TTTGACGTTCTTGAAGTCATTGGCTGCCGAGGGCTGAATCAAACTAACAACATCTGCTTC |
Microarray Signals from CUST_16796_PI426222305
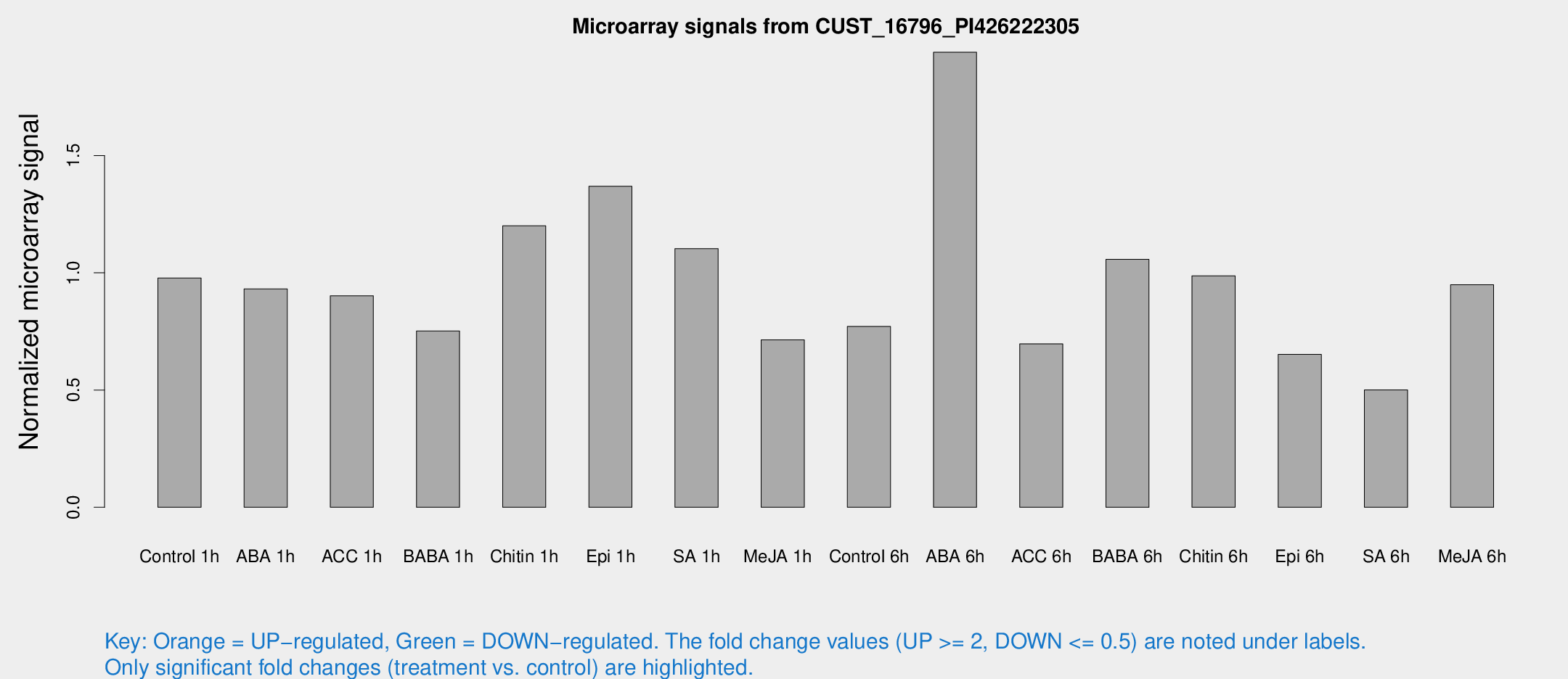
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 22.5335 | 6.00792 | 0.97765 | 0.354839 |
| ABA 1h | 19.2228 | 6.02915 | 0.931094 | 0.243098 |
| ACC 1h | 23.5489 | 8.03729 | 0.901773 | 0.408893 |
| BABA 1h | 15.9729 | 3.97752 | 0.751614 | 0.249766 |
| Chitin 1h | 24.8369 | 7.68881 | 1.20007 | 0.281034 |
| Epi 1h | 25.065 | 3.85618 | 1.36865 | 0.2107 |
| SA 1h | 23.9404 | 3.89364 | 1.10265 | 0.180423 |
| Me-JA 1h | 13.1271 | 3.83866 | 0.713951 | 0.24261 |
| Control 6h | 17.1272 | 3.90168 | 0.771217 | 0.200161 |
| ABA 6h | 43.9706 | 4.70411 | 1.94048 | 0.233491 |
| ACC 6h | 20.6722 | 7.54961 | 0.696812 | 0.359552 |
| BABA 6h | 26.4344 | 6.22528 | 1.05725 | 0.253059 |
| Chitin 6h | 22.6333 | 4.40341 | 0.987028 | 0.20042 |
| Epi 6h | 15.9199 | 4.48432 | 0.652134 | 0.190775 |
| SA 6h | 10.7763 | 4.00819 | 0.50076 | 0.199054 |
| Me-JA 6h | 20.0521 | 3.92425 | 0.948793 | 0.188435 |
Source Transcript PGSC0003DMT400069540 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G16090.1 | +1 | 0.0 | 572 | 303/412 (74%) | RING/U-box superfamily protein | chr3:5456513-5458694 FORWARD LENGTH=492 |
