Probe CUST_16784_PI426222305 - General Information
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16784_PI426222305 | JHI_St_60k_v1 | DMT400069541 | TTTGACGTTCTTGAAGTCATTGGCTGCCGAGGGCTGAATCAAACTAACAACATCTGCTTC |
All Microarray Probes Designed to Gene DMG402027033
| Probe ID | Chip name | Transcript ID | Probe Sequence |
|---|---|---|---|
| CUST_16784_PI426222305 | JHI_St_60k_v1 | DMT400069541 | TTTGACGTTCTTGAAGTCATTGGCTGCCGAGGGCTGAATCAAACTAACAACATCTGCTTC |
| CUST_16796_PI426222305 | JHI_St_60k_v1 | DMT400069540 | TTTGACGTTCTTGAAGTCATTGGCTGCCGAGGGCTGAATCAAACTAACAACATCTGCTTC |
Microarray Signals from CUST_16784_PI426222305
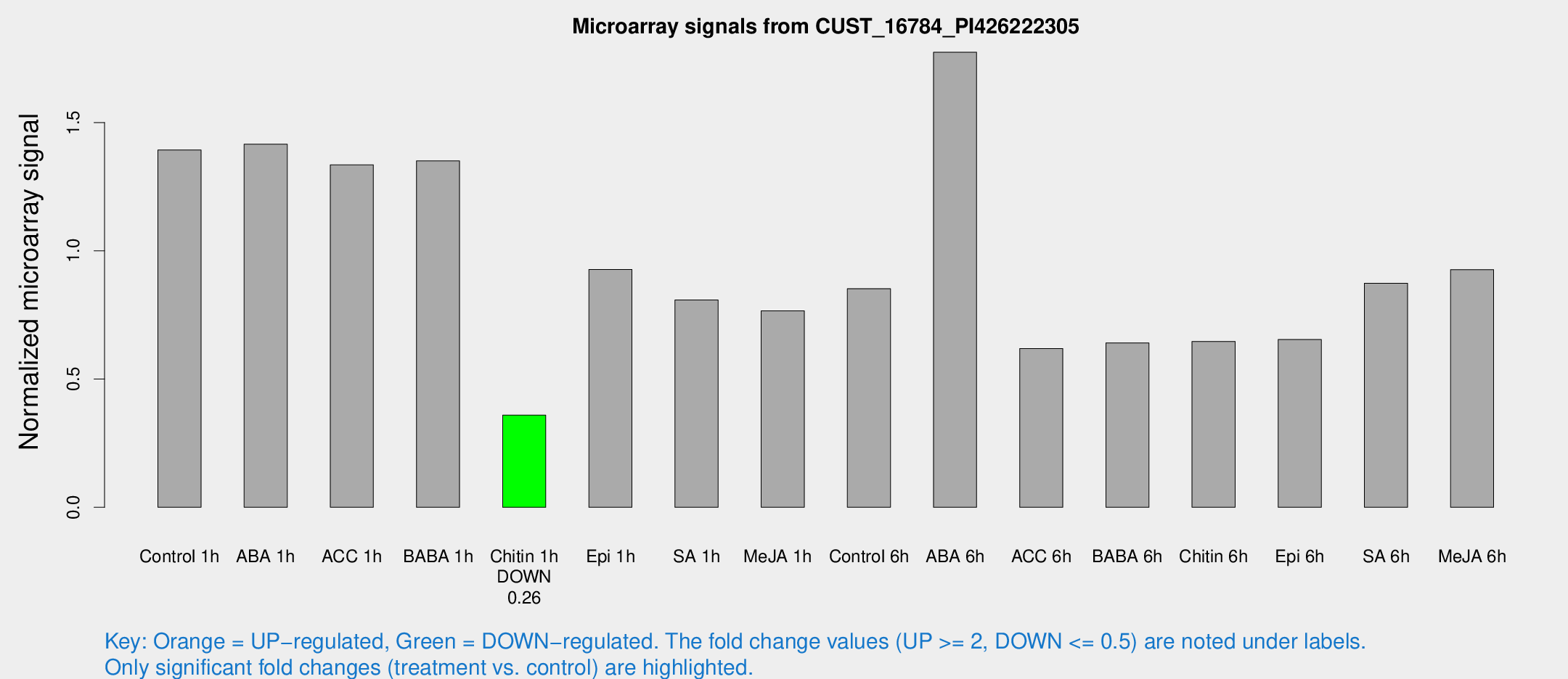
| Treatment | Raw signal | Raw Std Err | Normalized signal | Normalized Std Err |
|---|---|---|---|---|
| Control 1h | 27.4825 | 4.00285 | 1.39314 | 0.221664 |
| ABA 1h | 25.7694 | 5.33827 | 1.4155 | 0.273443 |
| ACC 1h | 27.5557 | 4.8443 | 1.33503 | 0.345816 |
| BABA 1h | 26.209 | 4.27638 | 1.35067 | 0.366021 |
| Chitin 1h | 6.3228 | 3.67325 | 0.359446 | 0.208416 |
| Epi 1h | 17.8985 | 5.42259 | 0.927438 | 0.449569 |
| SA 1h | 17.8168 | 4.93072 | 0.808168 | 0.241399 |
| Me-JA 1h | 13.4423 | 3.84204 | 0.766143 | 0.278087 |
| Control 6h | 20.2102 | 7.24392 | 0.852314 | 0.375654 |
| ABA 6h | 38.7009 | 7.84839 | 1.7742 | 0.336441 |
| ACC 6h | 15.6466 | 5.17331 | 0.619073 | 0.219893 |
| BABA 6h | 20.4632 | 12.5509 | 0.640925 | 0.497413 |
| Chitin 6h | 13.5632 | 4.21586 | 0.64682 | 0.206349 |
| Epi 6h | 15.4749 | 4.55359 | 0.654208 | 0.23779 |
| SA 6h | 20.2003 | 6.91978 | 0.873113 | 0.582656 |
| Me-JA 6h | 18.7708 | 3.88593 | 0.926784 | 0.216122 |
Source Transcript PGSC0003DMT400069541 - Homology to Model Species (BLASTX to E-value < 1e-50)
| Database | Link to BLAST Hit | Frame | E-value | Score | % Identity | Description |
|---|---|---|---|---|---|---|
| Tomato (ITAG) | None | - | - | - | - | - |
| TAIR PP10 | AT3G16090.1 | +3 | 4e-97 | 315 | 167/241 (69%) | RING/U-box superfamily protein | chr3:5456513-5458694 FORWARD LENGTH=492 |
